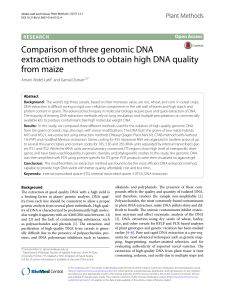
Fluorescence In Situ Hybridization (FISH) UNIT 22.4 As early as 1988 (Lichter et al., 1988), FISH was used to visualize labeled DNA probes hybridized to chromosome and interphase nuclei preparations. The improvement of cloned DNA sources, antibodies, fluorochromes, microscopy and imaging equipment, and software has permitted a variety of scientific investigations. This unit is divided into five parts—probe preparation, slide preparation, hybridization, post-hybridization washes, and interpretation. Probes are prepared by nick translation (see Basic Protocol 1) or degenerative oligonucleotide primer PCR (DOP-PCR; see Alternate Protocol 1) using hapten- or fluorochrome-labeled nucleotide. The amount of hapten label incorporated is quantified by dot blotting (see Support Protocol 1). Cytogenetic slide preparations are suitable for FISH (see Basic Protocol 2) if they are appropriately aged (see Support Protocol 2); these slides may be used even if they have previously been G-banded (see Alternate Protocol 2). In addition, slides made from paraffin-embedded tissues are suitable specimens for hybridization with DNA (see Alternate Protocol 3) or protein nucleic acid (PNA) probes (see Alternate Protocol 4). Hybridization conditions must be appropriate for both the sample and probe materials: cytogenetic slides can be hybridized with DNA (see Basic Protocol 3) and PNA (see Alternate Protocol 5) probes, as can paraffin sections (see Alternate Protocols 6 and 7). Post hybridization wash and detection conditions vary depending on the probe—indirectly labeled DNA probes (see Basic Protocol 4), directly labeled DNA probes (see Alternate Protocol 8 or 9), and peptide nucleic acid (PNA) probes (see Alternate Protocol 10). NOTE: Coplin jars containing solutions at temperatures other than room temperature should be prewarmed and maintained in a water bath at the specified temperature. LABELING DNA PROBES FOR FISH This section describes methods for labeling probes for fluorescence in situ hybridization (FISH) analysis. The first protocol (see Basic Protocol 1) describes DNA labeling by nick translation either indirectly with a hapten (biotin or digoxigenin) or directly with a fluorochrome (APPENDIX 1E) conjugated to dUTP. This method is appropriate for most DNAs listed in Table 22.4.1; however, smaller DNAs, such as cDNAs, may require labeling by PCR methods, which are also described (see Alternate Protocol 1). The choice of label, whether direct or indirect, is based on the type of DNA probe, its size, and the application. Small probe inserts (such as cDNAs) should be labeled with a hapten, enabling the option of signal amplification. Larger inserts can be both directly or indirectly labeled (Table 22.4.1). The probe’s signal strength can vary depending on the application and sequence specificity (Table 22.4.2). Furthermore, the choice of label or fluorochrome will also depend on the availability of conjugated antibodies, the nature of the experiment, and the availability of filters for the microscope utilized for visualization. A list of fluorochrome properties can be found in Table 22.4.3. For both labeling methods discussed here, assessment of probe labeling is determined by size, gel electrophoresis, and for indirectly labeled probes, dot blot analysis of incorporation (see Support Protocol 1). The labeled DNA is ethanol precipitated in the presence of excess unlabeled DNAs, which serve as carriers (sonicated salmon sperm DNA) and as suppressors of repetitive sequences (Cot-1 DNA). The final product is resuspended in a hybridization buffer and is then ready for use in FISH experiments (see Hybridization). The labeling procedures outlined can be applied to all species of DNA. Contributed by Jane Bayani and Jeremy A. Squire Current Protocols in Cell Biology (2004) 22.4.1-22.4.52 C 2004 by John Wiley & Sons, Inc. Copyright Cell Biology of Chromosomes and Nuclei 22.4.1 Supplement 23 Table 22.4.1 Sources of DNA Probes for FISH Analysisa Type Application Amplification of signal required Labeling methods applicable Indirect or direct labeling Plasmids (10–20 kb insert) Locus specific Yes Nick translation and PCR Indirect Cosmids (15–30 kb insert) Locus specific Yes Nick translation and PCR Indirect PACs (∼100 kb insert) Locus specific No Nick translation and PCR Both BACs (∼200 kb insert) Locus specific No Nick translation and PCR Both YACs (300–1.5 Mb insert) Locus specific No Nick translation and PCR Direct Genomic DNA from flow sorted chromosomes Chromosome paints No PCR labeling Both Genomic DNA from microdissected DNA Locus specific Yes PCR labeling Both a Shown are the most common types of DNAs used as probes for FISH analysis. Many of these DNAs can be labeled by nick translation (see Basic Protocol 1) or PCR (see Alternate Protocol 1). For probe inserts <2.0 kb, labeling by PCR is recommended. Table 22.4.2 Different Classes of Commonly Used Probes for FISH Analysisa Type Fluorescence intensity Typical applications Centromere probes Strong Ideal probe for the beginner to learn basic FISH techniques. Commonly used to enumerate chromosomal monosomies and trisomies, and for sex determination in transplantation studies. Subtelomere specific probes Moderate/weak Used for determining whether small terminal rearrangements near telomeres have taken place. Chromosome paints Strong/moderate Useful for identifying small marker chromosomal aberrations where a specific chromosome is suspected. Also useful in confirming SKY or MFISH findings. Translocation junction unique-sequence probes Moderate Used for detecting the presence of specific translocations in interphase cells or in metaphase spreads. Microdeletion unique-sequence probes Moderate Identification of small submicroscopic deletions using metaphase preparations. Probes detecting gene amplification Strong For detecting oncogene copy number increases (gene amplification) in interphase and metaphase cells a Shown are the most common types of commercially available probes used in FISH experiments and their typical applications. Signal strength will vary depending on the size and application of the probe as well as the target DNA specimen. Another choice for FISH analysis is a peptide nucleic acid (PNA) probe. These probes also bind DNA in a sequence-specific manner but are created by oligonucleotide synthesis. The fabrication of PNA probes is not easily accomplished in a typical molecular cytogenetic laboratory, so they are currently only available commercially (see Internet Resources). BASIC PROTOCOL 1 Fluorescence In Situ Hybridization (FISH) Labeling FISH Probes by Nick Translation This protocol, adapted from Beatty et al. (2002), outlines the basic steps for either directly or indirectly labeling DNA probes for FISH analysis. Once labeled probes are ethanol precipitated, they are ready for final preparation prior to hybridization to the target specimen. Materials DNA probe (i.e., cosmid, plasmid, PAC, BAC, or YAC; Table 22.4.1) 10× nick translation buffer (see recipe) 22.4.2 Supplement 23 Current Protocols in Cell Biology Table 22.4.3 Fluorescent Properties of Labels and DNA Stains Used for FISHa Label Excitation (nm) Emission (nm) Digoxigenin NA NA Biotin NA NA Alexa 488c 490 520 c 525 550 c 555 570 c Alexa 594 590 615 Amino-methyl coumarin 399 445 Cascade Blue Haptens b Fluorochromes Alexa 532 Alexa 546 400 420 d 489 506 d 550 570 d 649 670 d Cyanine 7 743 767 Fluorescein isothiocyanate (FITC) 495 523 Rhodamine B Cyanine 2 Cyanine 3 Cyanine 5 560 580 e 433 480 e 655 675 497 524 530 555 Spectrum Orange 559 588 Spectrum Rede 587 612 Tetramethylrhodamine isothiocyanate (TRITC) 550 570 Texas Red 595 610 Chromomycin A3 430 570 4 ,6-Diamindine-2-phenylindole (DAPI) 538 461 Ethidium bromide 518 615 Hoechst 33258 (bis-benzimide) 352 461 Propidium bromide 535 617 Spectrum Aqua Spectrum FRed e Spectrum Green e Spectrum Gold e DNA stains a Shown are the commonly used fluorochromes and DNA stains used in FISH analysis. Of these, DAPI is the most widely used. b Haptens, such as digoxigenin and biotin are detected with antibodies conjugated to one of the fluorochromes listed. c The Alexa family of fluorochromes can be purchased from Molecular Probes. d The Cyanine family of fluorochromes can be purchased from Amersham Biosciences. e The Spectrum family of fluorochromes can be purchased from Vysis. DNase I dilution buffer (see recipe) 1 mM dNTP mixture (see recipe) Fluorochrome/dTTP or hapten/dTTP mixture (see recipes) 3 mg/ml DNase I (see recipe) 10 U/µl E. coli DNA polymerase I (Roche) 5 × loading dye (see recipe) Cell Biology of Chromosomes and Nuclei 22.4.3 Current Protocols in Cell Biology Supplement 23 2% (w/v) agarose gel (see recipe) 100-bp DNA ladder Sonicated salmon sperm DNA standards (i.e., 12.5, 25.0, and 500 ng/µl; see recipe) 1× TBE buffer (see APPENDIX 2A) 300 mM EDTA (APPENDIX 2A) 10 mg/ml sonicated salmon sperm DNA (Invitrogen) 1 µg/µl human or mouse Cot-1 DNA (Invitrogen) 3 M sodium acetate (APPENDIX 2A) 100% ethanol 70% ethanol, cold Hybridization buffer (see recipe) Water bath or PCR machine 0.5-ml PCR tube (optional) Additional reagents and equipment for agarose gel electrophoresis (APPENDIX 3A) and determination of hapten incorporation by dot blot analysis (see Support Protocol 1) Prepare equipment and reagents 1. Set water bath or PCR machine to 15◦ C. 2. Put DNA probe and all labeling reagents (i.e., 10× nick translation buffer, DNase I dilution buffer, 1 mM dNTP mixture, and fluorochrome/dTTP or hapten/dTTP mixture) on ice. 3. Prepare 10× DNase I solution (1.0 ml total) by combining 1.0 µl of 3 mg/ml DNase I and 999.0 µl DNase I dilution buffer. Place on ice. Perform labeling reaction 4. For each DNA to be labeled, prepare the following mixture in a 1.5-ml microcentrifuge or 0.5-ml PCR tube, adding the 10× DNase I solution and E. coli polymerase I last: 2 µg DNA probe 10.0 µl 10× nick translation buffer 5.0 µl 1 mM dNTP mixture (i.e., dATP, dCTP, dGTP) 4.0 µl fluorochrome/dTTP or hapten/dTTP mixture 10.0 µl 10× DNase I solution 2.5 µl E. coli DNA polymerase. Adjust the volume to 100 µl with water. The amount of DNase I solution added will vary (see Commentary). A probe prepared using hapten/dTTP is indirectly labeled, while one prepared with fluorochrome/dTTP is directly labeled. 5. Incubate 90 to 120 min at 15◦ C (see Table 22.4.22 and Critical Parameters). 6. After incubation, place tubes on ice. Determine size of products from labeling reaction 7. Transfer 10 µl labeling reaction to a fresh tube and mix with 4 µl of 5× loading dye. Load onto a 2% agarose gel (APPENDIX 3A) along with a 100-bp DNA marker and 10 µl of each salmon sperm concentration standard. Fluorescence In Situ Hybridization (FISH) The salmon sperm standards are used to help determine the amount of labeled DNA probe, as well as to act as a molecular-size marker. 8. Electrophorese at 100 V for ∼20 min or until the dye front is two thirds of the way through the gel. 22.4.4 Supplement 23 Current Protocols in Cell Biology Figure 22.4.1 Nick translated DNA fragments electrophoresed on a 2% agarose gel. A volume of 10.0 µl labeled product was loaded. The fragment sizes range from 200 to 500 bp as determined from the molecular marker. The estimated concentration of the probe is 200 ng in 10 µl, yielding ∼20 ng/µl. The final fragment sizes should be between 200 and 500 bp, with ∼200 ng labeled DNA present in the gel, assuming the original amount of labeled probe was 2 µg. This can be ascertained using the sonicated salmon sperm concentration standards (Figure 22.4.1). 9. If the fragment sizes are too large, return the labeling reaction to 15◦ C for 20 to 30 min (or time as required) and reassess the size of the fragments (i.e., repeat steps 5 to 8). It may be necessary to spike the labeling reaction with additional DNase I solution and E. coli DNA polymerase I. If the fragments are too small, labeling (step 4) will need to be repeated with an adjustment in the amount of DNase I added to the labeling reaction. If there is no significant change in fragment size following further digestion, the investigator should start again and review Table 22.4.22 for possible reasons of insufficient digestion. Determine amount of labeled probe 10. Once the proper labeling size has been achieved, add 10 µl of 300 mM EDTA to the labeling reaction to stop the action of the enzymes. 11. Estimate the amount of labeled probe on the gel using the salmon sperm standards as a guide. Calculate the amount of labeled DNA remaining in the tube. Be sure to consider the volumes removed from the starting volume for loading onto the gel(s) when calculating the amount of labeled DNA left in the tube. Based on the concentration standards, the labeled probe in Figure 22.4.1 appears to be somewhere between 125 ng/10 µl (1.25 ng/µl) and 250 ng/10 µl (25.0 ng/µl). A conservative estimate is 200 ng/10 µl (20 ng/µl). Thus, from an original 100 µl starting volume, 10 µl was removed for a gel, leaving 90 µl. If the estimated concentration is 20 ng/µl, the remaining labeled probe is 1800 ng (20 ng/µl × 90 µl = 1800 ng). Cell Biology of Chromosomes and Nuclei 22.4.5 Current Protocols in Cell Biology Supplement 23 Add carrier and supressor of repetitive sequences 12. For each 1 µg labeled DNA in the tube, add 5 µl of 10 mg/ml sonicated salmon sperm DNA (50 µg). For example, for 1.8 µg DNA, add ∼100 µg sonicated salmon sperm DNA. 13. For each microgram labeled DNA in the tube, add 5 to 10 µg human or mouse Cot-1 DNA. For the example given above (1.8 µg labeled DNA), ∼20 µg Cot-1 DNA is required The amount of Cot-1 used is dependent on the number and extent of repeat elements in the DNA insert used as the probe. Generally, cDNA probes will have fewer repeats than genomic probes, so they will require relatively less Cot-1 suppression. BAC and YAC probes may need more suppression. Determine incorporation 14. If the DNA probe has been labeled with hapten, check incorporation by dot blot analysis (see Support Protocol 1). Ethanol precipitate DNA 15. Add 0.1 vol. of 3 M sodium acetate followed by 2.5 vol of 100% ethanol. 16. Incubate overnight at −20◦ C or 2 to 3 hr at −80◦ C. The recovery of DNA during the precipitation process can be increased by lengthening the time at −20◦ C or −80◦ C. 17. Microcentrifuge 20 min at 13,000 rpm, 4◦ C. Resuspend DNA 18. Carefully remove the supernatant and wash the pellet with cold 70% ethanol. 19. Microcentrifuge 20 min at 13,000 rpm, 4◦ C. 20. Remove the ethanol and allow the pellet to air dry. 21. Resuspend the pellet in hybridization buffer to a final concentration of 10 ng/µl. Store labeled probe at −20◦ C up to several months (directly labeled probe) or several years (indirectly labeled probe). The final concentration should be based on the labeled DNA content, not the total DNA content, which includes the unlabeled Cot-1 and salmon sperm DNA. Thus, for a labeled DNA amount of 1.8 µg, 180 µl hybridization buffer should be added to reach a final concentration of 10 ng/µl. The labeled probe can now be used for FISH experiments. The amount of labeled probe used will depend on the area of the slide to be hybridized. For example, 10 µl is sufficient to cover a 22 × 22–mm coverslip, 20 µl is sufficient to cover a 22 × 30–mm coverslip, and 30 µl is sufficient to cover a 22 × 50–mm coverslip. ALTERNATE PROTOCOL 1 Labeling Probes for FISH by Degenerative Oligonucleotide Primer Polymerase Chain Reaction (DOP-PCR) This protocol describes direct and indirect labeling procedures using PCR labeling. The user has various choices in the primer sets. These may be specific primers for a region, specific primers for vector sequences, universal primers, or DOP primers. Additional Materials (also see Basic Protocol 1) Fluorescence In Situ Hybridization (FISH) DNA template (Table 22.4.1) 10× PCR buffer: 100 mM Tris·Cl, pH 8.3 (APPENDIX 2A)/500 mM KCl (store up to several months at −20◦ C) 22.4.6 Supplement 23 Current Protocols in Cell Biology 2 mM dNTP mixture (i.e., dATP, dCTP, dGTP; see recipe) 5 µM each primers 1 and 2 (APPENDIX 3F) 50 mM MgCl2 5 to 10 U/µl Taq DNA polymerase Water, sterile Mineral oil, sterile 1. For each labeling reaction, mix the following: 1 to ng DNA template 5.0 µl 10× PCR Buffer 4.0 µl 2 mM dNTP 2.0 µl 1 mM fluorochrome/dTTP or hapten/dTTP 1.5 µl 5 µM primer 1 1.5 µl 5 µM primer 2 1.5 µl 50 mM MgCl2 0.5 µl 5–10 U/µl Taq DNA polymerase. Adjust final volume to 50.0 µl with sterile water. Overlay the mixture with sterile mineral oil and put on ice until ready to place in the thermocycler. The 10× PCR buffer is often supplied with the enzyme. 2. Place in a thermocycler and program the following steps: Initial step: 35 cycles: Final step: Hold: 5 min 30 sec 30 sec 90 sec 5 min indefinitely 93◦ C 94◦ C 55◦ C 72◦ C 72◦ C 4◦ C. (denaturation) (denaturation) (annealing) (extension) (extension) 3. Mix 5 µl labeled reaction with 1 µl of 5× loading dye and run on a 2% (w/v) agarose gel with molecular weight markers and sonicated salmon sperm DNA concentration standards as described (see Basic Protocol 1, steps 7 and 8). The final PCR product should fall between 200 and 500 bp. 4. Calculate the amount of labeled DNA remaining in the tube, add carrier and supressor DNA, precipitate, and resuspend in hybridization buffer as described (see Basic Protocol 1, steps 11 to 20). 5. If the DNA probe has been labeled with hapten, check incorporation by dot blot analysis (see Support Protocol 1). Determination of Hapten Incorporation by Dot Blot Analysis Gel electrophoresis of the labeled DNA FISH probe allows the investigator to determine whether the DNA has been nicked (see Basic Protocol 1) or amplified (see Alternate Protocol 1) to the appropriate sizes. It does not, however, allow determination of whether the hapten has been incorporated into the DNA. Using a dot blot assay, the degree of hapten incorporation can be assessed to determine the efficiency of the labeling protocol and the activity of the DNA polymerase I or Taq DNA polymerase. This method is useful in determining whether the lack of FISH signal is due to a poorly labeled probe or to hybridization or slide preparation factors. This method involves spotting labeled product onto a nylon filter along with a control. The labeled DNA is detected by immunolabeling and colorimetric visualization. SUPPORT PROTOCOL 1 Cell Biology of Chromosomes and Nuclei 22.4.7 Current Protocols in Cell Biology Supplement 23 Materials 100 mM Tris·Cl (pH 7.5)/15 mM NaCl (APPENDIX 2A) Indirectly labeled probe (i.e., with biotin or digoxigenin; Table 22.4.3) Control DNA labeled with biotin or digoxigenin (i.e., labeled DNA probe known to produce good signal strength) 0.5% (w/v) BSA in 100 mM Tris·Cl, pH 7.5 (APPENDIX 2A)/15 mM NaCl (store up to 6 months at 4◦ C) AP-labeled antibody mixture (see recipe) 100 mM Tris·Cl (pH 9.5)/100 mM NaCl/50 mM MgCl2 (APPENDIX 2A) NBT/BCIP (see recipe) Charged nylon membrane (∼5 × 5 cm per sample) Filter paper 37◦ C dry oven Rotating platform Prepare and load nylon membrane 1. Cut a charged nylon membrane to a size appropriate for the number of samples. 2. Soak the membrane 5 min in 100 mM Tris·Cl (pH 7.5)/15 mM NaCl, room temperature. Blot the membrane with filter paper. 3. Pipet aliquots of indirectly labeled probe and control DNA labeled with biotin or digoxigenin in a dilution series (e.g., 200, 100, 50 ng) onto the membrane, leaving ample space between spots. Incubate 5 to 10 min, room temperature. Block and label membrane 4. Incubate the membrane 1 min in 100 mM Tris·Cl (pH 7.5)/15 mM NaCl, making sure it is saturated. 5. Incubate the membrane 30 min in 0.5% (w/v) BSA in 100 mM Tris·Cl (pH 7.5)/15 mM NaCl at room temperature. 6. Transfer the membrane to an alkaline phosphatase–labeled antibody mixture in a plastic container and incubate 30 min in a 37◦ C dry oven on a rotating platform, making sure the membrane is completely saturated. Develop dot blot 7. Remove the membrane and wash 15 min with 100 mM Tris·Cl (pH 7.5)/15 mM NaCl. 8. Remove the membrane and wash 2 min with 100 mM Tris·Cl (pH 9.5)/100 mM NaCl/50 mM MgCl2 , room temperature. 9. Remove the membrane and incubate 5 to 10 min in NBT/BCIP solution under dimmed lighting until the blot develops. 10. Wash the membrane with water and air dry. The membrane can be stored at room temperature. Over time the intensity of the chemical stain will fade. Typical results are shown in Figure 22.4.2. TARGET SLIDE PREPARATION Fluorescence In Situ Hybridization (FISH) Slide quality can greatly influence the success of FISH assays. When slide preparations are less than optimal, treatment with a protease can decrease the risk of high background and increase access of the probe to the DNA target by removing protein barriers to allow efficient hybridization. FISH carried out on paraffin sections requires more aggressive 22.4.8 Supplement 23 Current Protocols in Cell Biology Figure 22.4.2 Dot-blot analysis of biotin incorporation in probes shown in Figure 21.4.1. Compared to control labeled DNA, the labeled probes appear to have incorporated biotin well, with probe 4 showing the greatest incorporation compared to probe 1. The concentration of each probe was estimated at 20 ng/µl; however, it is evident that the labeling efficiency was greater in some probes over others. protease treatment and assessment. Following optimal slide pretreatment, the specimen is subjected to denaturation, causing double-stranded DNA to become single stranded. This will permit the efficient hybridization of the denatured probe to the DNA target on the slide. Preparation of Cytogenetic Specimens for FISH The following protocol describes the pretreatment of cytogenetic slides (UNIT 22.2) for FISH analysis with DNA probes (in-house or commercial) or commercially obtained PNA probes. In this protocol, pepsin is recommended for use as the protease; however, proteinase K is also often used. The concentration and conditions for proteinase K treatment are also noted in the protocol. The treatment with pepsin will vary depending on the extent of digestion required on the slide. In some cases, pepsin treatment is not needed. BASIC PROTOCOL 2 Materials Cytogenetic slide preparation (UNIT 22.2): age naturally at least 2 days at room temperature or artificially (see Support Protocol 2) 10% (w/v) pepsin—100 mg pepsin powder (Sigma) in 1.0 ml H2 O; store in 20-µl aliquots up to several months at −20◦ C—and 0.01 M HCl, 37◦ C or 14 mg/ml proteinase K (Roche Diagnostics) and proteinase K buffer—i.e., 20 mM Tris·Cl, pH 7 (APPENDIX 2A)/0.2 mM CaCl; store up to several months at room temperature 1× PBS (APPENDIX 2A) 70% formamide/2× SSC (pH 7.0), 72◦ C (see UNIT 18.6 for SSC): prepare fresh 70% ethanol, ice-cold (for DNA probes) Cell Biology of Chromosomes and Nuclei 22.4.9 Current Protocols in Cell Biology Supplement 23 Table 22.4.4 Coplin Jars Needed for Preparation of Cytogenic Specimens for FISHa No. Coplin jars 1 Contents Temperature 70% ethanol 1 70% ethanol 1 or 2b b 1 or 2 Room temperature 4◦ C b 80% ethanol Room temperature 100% ethanol Room temperature b 72◦ C 1 70% formamide in 2× SSC 1 1× PBS Room temperature 1 Pepsin/0.01 HCl or proteinase K/buffer 37◦ C or room temperature (respectively) a See Basic Protocol 2. b For DNA probes only. Phase contrast microscope (UNIT 4.1) Coplin jars Additional reagents and equipment for hybridization (see Basic Protocol 3 and Alternate Protocol 5) CAUTION: Formamide is a carcinogen and should be handled with care. Discard according to biohazard rules of the institution. NOTE: A list of all Coplin jars used in this protocol is given in Table 22.4.4. Detect and remove cytoplasmic contamination by protease treatment 1. Using a phase-contrast microscope, determine the extent of cytoplasmic residue on the cytogenetic slide preparation. If there is no cytoplasm, proceed to step 6. 2a. For pepsin digestion: Add 10 to 15 µl of 10% (w/v) pepsin to 50 ml warm 0.01 M HCl in a Coplin jar. Incubate the slides in the protease 5 min at 37◦ C. 2b. For proteinase K digestion: Dilute 14 mg/ml proteinase K to a final concentration of 0.1 µg/ml in proteinase K buffer in a Coplin jar. Incubate the slides in the protease 6.5 min at room temperature. Remove protease and determine efficacy 3. Wash the slide 5 min at room temperature in a Coplin jar containing 1× PBS. 4. Pass the slide through a dehydrating series of 70%, 80%, and 100% ethanol in Coplin jars for 5 min each. Allow to air dry after the final ethanol treatment (∼5 min). 5. View the slide using phase-contrast microscopy to determine the extent of protein digestion. If necessary, repeat the protease step. Prepare slides for hybridization 6a. For PNA probes: Proceed to hybridization (see Alternate Protocol 5). 6b. For DNA probes: Denature the slide for the recommended time (see Table 22.4.5) in 70% formamide/2× SSC (pH 7.0), 72◦ C. Fluorescence In Situ Hybridization (FISH) The time will vary according to the age and quality of the slide. 7b. Promptly place the slide into a Coplin jar containing ice-cold 70% ethanol for 5 min. Pass the slides through a dehydration series of 80% and 100% ethanol for 5 min each. 22.4.10 Supplement 23 Current Protocols in Cell Biology Table 22.4.5 Formamide Denaturation Guide for Cytogenetic Specimensa Slide criteria Suggested denaturation timeb Fresh slide stored 1–2 days at room temperature 60–90 sec Fresh slide artificially aged 1–2 days 60–90 sec Aged slide stored 1–2 weeks at room temperature 1.5–2 min Aged slide stored 2–4 weeks at room temperature 2 min Aged slides stored 1–2 months 2–3 min Previously G-banded slides >1-week old 30–40 sec Previously G-banded slides <1-week old 20–30 sec a Shown are general guidelines for denaturation of slides of varying ages and conditions. The investigator should pay attention to the slide quality and monitor the changes in denaturation conditions as the slides age or are processed. Each laboratory will possess different environmental conditions as well as slide making procedures, which will affect the denaturation procedures significantly. b Denaturation conditions are 72◦ C in 70% formamide/2× SSC. 8b. Air-dry the slide after the final ethanol treatment and proceed to hybridization (see Basic Protocol 3). Artificial Aging of Cytogenetic Slide Preparations for FISH The chromosomes on freshly prepared slides are often too fragile for immediate use— i.e., the high temperatures used during the slide denaturation procedure can damage the DNA, making it less optimal for hybridization. Such slides require aging, which can be achieved by allowing them to naturally age a few days at ambient temperature; however, an experiment must occasionally be performed immediately. The protocol below outlines a method for artificially aging freshly prepared slides so that FISH results are available within 12 to 24 hr after preparation from the cytogenetic suspension. This method may also be used when the relative humidity in the laboratory is high due to local weather conditions. SUPPORT PROTOCOL 2 To age, incubate a freshly prepared cytogenetic slide (UNIT 22.2) at least 1 hr (up to 3 hr) in a Coplin jar containing 2× SSC (UNIT 18.6), 37◦ C. Remove the slide and pass through a dehydration series of 70%, 80%, and 100% ethanol for 5 min each, and air dry (5 to 10 min). Proceed with enzyme digestion if required. Note that a list of all Coplin jars used in this protocol is given in Table 22.4.6. Preparation of Previously G-Banded Cytogenetic Specimens for FISH The following method outlines the steps involved in pretreating a previously Giemsa (G)-banded cytogenetic slide (UNIT 22.3) for subsequent use in FISH analysis. The slides must be completely free of any oils and must also be destained. Since the slide has already been treated with a protease, it will require no additional protease digestion; however, stripping the proteins, which are protective to the DNA, will cause it to become much more sensitive to degradation during the denaturation process. The ability to re-use a previously banded slide for FISH will be dependent on the age of the slide, the degree of trypsin digestion during the banding, and whether residual immersion oils or mounting buffers have degraded the DNA. ALTERNATE PROTOCOL 2 Materials Banded cytogenetic slide specimen (UNIT 22.3) Xylene Cell Biology of Chromosomes and Nuclei 22.4.11 Current Protocols in Cell Biology Supplement 23 Table 22.4.6 Coplin Jars Needed for Artificial Aging of Cytogenetic Slide Preparations for FISHa No. Coplin jars Contents Temperature 1 70% ethanol Room temperature 1 80% ethanol Room temperature 1 100% ethanol Room temperature 1 2× SSC 37◦ C a See Support Protocol 2. Table 22.4.7 Coplin Jars Needed for Preparation of Previously G-Banded Cytogenetic Specimens for FISHa No. Coplin jars Contents Temperature 1 70% ethanol 4◦ C 1 70% ethanol Room temperature 2 80% ethanol Room temperature 2 100% ethanol Room temperature 1 70% formamide/2× SSC, pH 7.0 72◦ C 1 Methanol Room temperature 1 b Xylene Room temperature a See Alternate Protocol 2. b CAUTION: Maintain in a fume hood. Methanol 70%, 80%, and 100% ethanol 70% formamide in 2× SSC (pH 7.0; UNIT 18.6), 72◦ C: prepare fresh 70% ethanol, ice-cold Coplin jars NOTE: A list of all Coplin jars used in this protocol is given in Table 22.4.7. 1. Remove residual oils from previously banded cytogenic slide specimen by incubating 5 min in a Coplin jar containing xylene under a vented chemical hood. 2. Destain by incubating the slide ∼5 to 10 min in a Coplin jar containing roomtemperature methanol. 3. Pass the slide through a dehydrating series of 70%, 80%, and 100% ethanol for 5 min each. Allow the slide to air dry (5 to 10 min) after the final ethanol treatment. 4. Denature slide 20 to 30 sec in 70% formamide/2× SSC (pH 7.0), 72◦ C. The time will vary according to the age and quality of the slide (Table 22.4.5). 5. Promptly place the slide into a Coplin jar containing ice-cold 70% ethanol for 5 min following denaturation. Fluorescence In Situ Hybridization (FISH) 6. Proceed through a dehydration series of 80% and 100% ethanol for 5 min each. Airdry the slide after the final ethanol treatment and proceed with hybridization (see Hybridization). 22.4.12 Supplement 23 Current Protocols in Cell Biology Preparation of Paraffin-Embedded Specimens for Hybridization with DNA Probes FISH analysis on paraffin sections is becoming increasingly important in both diagnostic and research laboratories. This protocol outlines the steps involved in pretreating paraffin sections for FISH. While the preparation of paraffin-embedded specimens can be modified, the most critical component is the quality of the starting section (see Commentary). Different histopathology laboratories will have various methods of tissue fixation which should be taken into consideration when assessing the success of an experiment. ALTERNATE PROTOCOL 3 Materials 5- to 10-µM paraffin sections on silanized slides Xylene 100% ethanol 0.5% (w/v) pepsin in 0.85% (w/v) NaCl (pH 1.5), 45◦ C (see recipe) 2× SSC, pH 7.0 (UNIT 18.6) Propidium iodide (PI) or DAPI in antifade (see recipes) 70%, 80% and 100% ethanol Coplin jars 45◦ C water bath Fluorescence microscope (UNIT 4.2) with a FITC, and PI or DAPI filter set NOTE: A list of all Coplin jars used in this protocol is given in Table 22.4.8. Remove wax 1. Add 5- to 10-µM paraffin sections on silanized slide to a Coplin jar containing xylene. Gently agitate 5 min at room temperature. Transfer slides to a second Coplin jar containing fresh xylene for an additional 5 min with gentle agitation. 2. Transfer the slide to a Coplin jar containing 100% ethanol. Soak 5 min, then transfer slides to fresh 100% ethanol and soak another 5 min. Agitate 2 to 3 times during each 5-min period. 3. Remove the slide from ethanol and allow to air dry (5 to 10 min). Perform protease digestion and counterstain 4. Place slide in a Coplin jar containing 0.5% (w/v) pepsin in 0.85% NaCl, 45◦ C. Incubate 15 to 20 min in a 45◦ C water bath (see Table 22.4.5). 5. Rinse the slide 20 to 30 sec in a Coplin jar containing 2× SSC, pH 7.0. Table 22.4.8 Coplin Jars Needed for Preparation of Paraffin-Embedded Specimens for Hybridization with DNA Probesa No. Coplin jars Contents Temperature 1 70% ethanol Room temperature 1 80% ethanol Room temperature 3 100% ethanol Room temperature 1 0.5% pepsin in 0.85% NaCl 45◦ C 4 2× SSC, pH 7.0 Room temperature 2 b Room temperature Xylene a See Alternate Protocol 3. b CAUTION: Maintain in a fume hood. Cell Biology of Chromosomes and Nuclei 22.4.13 Current Protocols in Cell Biology Supplement 23 Table 22.4.9 Troubleshooting Guide for Pretreatment of Paraffin-Embedded Sections Problem Cause Green autofluorescence Under-digestion with upon inspection/or poor pepsin solution uptake of counterstain Solution Remove coverslip and wash with 2× SSC as described (see Alternate Protocol 3, step 5). Return the slide to pepsin digestion buffer (see Alternate Protocol 3, step 4). The time required for subsequent protease treatment will be dependent on the existing degree of digestion. Should repeated attempts at digestion fail, consider another enzyme such as proteinase K (use at 10 mg/ml in 2× SSC for 20 min at 37◦ C) in place of the pepsin buffer (see Alternate Protocol 3, step 4) or in combination with the pepsin buffer. Lifting of tissue from slide Holes in tissue or “ghosty” appearance Loosening by protease treatment Occasionally the protease treatment will loosen the tissue from the slide. Try to work carefully such that the tissue is relatively undisturbed. Glass slides not coated with silane Associated with this is the use of charged or silanated slides. Many pathology labs will use coated slides. Check if the sections were mounted onto such slides. If not, try to obtain sections on these slides. Over-digestion Over-digestion will be evident from the degradation of the tissue. Start again and decrease the tissue treatment. Figure 22.4.3 Digestion of paraffin-embedded specimen for FISH. (A) Under-digestion is indicated by weak uptake of the DAPI stain. (B) Increased digestion permits better access to the DNA and increase uptake of DAPI. This black and white facsimile of the figure is intended only as a placeholder; for full-color version of figure, see color plates. 6. Apply 10 µl PI or DAPI in antifade to counter stain and coverslip. The choice between PI or DAPI depends on the label color of the probe being analyzed. Fluorescence In Situ Hybridization (FISH) 7. View the slide using a fluorescence microscope with FITC, and PI or DAPI filter set. Evaluate the tissue sections for under-digestion, appropriate digestion, or overdigestion using the guidelines in Table 22.4.9 and Figure 22.4.3. 22.4.14 Supplement 23 Current Protocols in Cell Biology For certain types of tissue additional troubleshooting steps may also be necessary (see Troubleshooting). 8. Carefully remove the coverslip and rinse slide three times in separate Coplin jars containing 2× SSC, pH 7.0, for 5 min each, agitating 5 to 10 sec in each rinse. 9. Dehydrate slide in a series of 70%, 80%, and 100% ethanol washes for 1 min each. 10. Allow the slide to air dry. Proceed to hybridization with labeled DNA probes. Preparation of Paraffin-Embedded Specimens for Hybridization with PNA Probes PNA probes are also applicable to paraffin sections. Like pretreatment for hybridization with DNA probes (see Alternate Protocol 3), pretreatment for hybridization with PNA probes requires some minor modifications as described below. ALTERNATE PROTOCOL 4 Materials 5- to 10-µM paraffin sections on silanized slides Xylene 100% ethanol 100 µg/ml RNase I (see recipe) 2× SSC (UNIT 18.6) 1 M sodium thiocyanate, 80◦ C 5 mg/ml pepsin in 0.85% (w/v) NaCl, 45◦ C (see recipe) 0.1 M triethanolamine, pH 8.0 Acetic anhydride (Sigma) 1× PBS (APPENDIX 2A) 70%, 80%, and 100% ethanol 45◦ C hot plate or slide warmer Coplin jars 37◦ C oven NOTE: A list of all Coplin jars used in this protocol is given in Table 22.4.10. Remove wax 1. Preheat 5- to 10-µm paraffin sections on silanized slide on a 45◦ C slide warmer or hot plate for ∼30 min (i.e., until the wax melts). Table 22.4.10 Coplin Jars Needed for Preparation of Paraffin-Embedded Specimens for Hybridization with PNA Probesa No. Coplin jars Contents Temperature 1 70% ethanol Room temperature 1 80% ethanol Room temperature 4 100% ethanol Room temperature 2 1× PBS Room temperature 1 5 mg/ml pepsin in 0.85% NaCl 45◦ C 1 1 M sodium thiocyanate 80◦ C 5 2× SSC Room temperature 1 Triethanolamine, pH 8.0 Room temperature 2 Water Room temperature 3 b Xylene a See Alternate Protocol 4. b CAUTION: Maintain in a fume hood. Room temperature Cell Biology of Chromosomes and Nuclei 22.4.15 Current Protocols in Cell Biology Supplement 23 2. Dewax by transferring the slide to a Coplin jar containing 50 ml xylene and incubating 5 to 10 min at room temperature. Repeat twice with fresh xylene. Time for dewaxing is dependent on many factors (e.g., tissue section thickness, surface area, type of tissue). 3. Remove xylene by soaking slide 5 to 10 min in 100% ethanol. Repeat twice with fresh ethanol. 4. Allow slides to air dry (5 to 10 min). The tissue should turn white at this point. Treat with RNase, SSC, and sodium thiocyanate 5. Add 40 µl of 100 µg/ml RNase I to the slide. Cover with a coverslip and incubate 1 hr at 37◦ C. 6. Wash twice in Coplin jars containing fresh 2× SSC for 5 min each time. 7. Incubate slide 8 min in a Coplin jar containing 1 M sodium thiocyanate, 80◦ C. 8. Rinse twice in Coplin jars containing fresh distilled water for 1 min each. Digest with protease 9. Incubate 7 to 9 min at 45◦ C in a Coplin jar containing 5 mg/ml pepsin in 0.85% NaCl, pH 1.5. 10. Wash twice for 1 min each in Coplin jars containing fresh 2× SSC. Acetylate 11. Start acetylation by placing slide in a Coplin jar containing 0.1 M triethanolamine, pH 8.0. 12. While stirring gently, slowly add 125 µl acetic anhydride to give a final concentration of 0.25% (v/v). Incubate 10 min at room temperature. The acetylation procedure is carried out to reduce the electrostatic binding of probe to positive charges on the tissue, thereby reducing the background. Wash and dehydrate 13. Rinse 5 min in a Coplin jar containing 1× PBS. Repeat the rinse using a fresh Coplin jar of PBS. Washes from this point forward can be reduced in duration if adherence of tissue to the slide is poor. 14. Rinse 5 min in a fresh Coplin jar containing 2× SSC. 15. Dehydrate through a series of 70%, 80%, and 100% ethanol and allow to air dry. 16. Proceed to hybridization with PNA probes (see Alternate Protocol 7). HYBRIDIZATION Fluorescence In Situ Hybridization (FISH) In the following section, hybridization of the probe to the slide is described. The prepared DNA probe is denatured to a single-stranded state in the presence of denaturing buffer. Depending on the probe type, there may be a preannealing step. The denatured probe is applied to the denatured cytogenetic specimen and allowed to hybridize overnight. Additionally, this section also describes the hybridization conditions for paraffin sections, where the slide and probe are simultaneously denatured. If commercial DNA probes are being used, the manufacturer’s instructions for probe denaturation are followed and the hybridization conditions are adjusted accordingly. If a PNA probe is utilized, the probe 22.4.16 Supplement 23 Current Protocols in Cell Biology is applied to the slide, and together the probe and target are denatured and permitted to hybridize for at least 1 hr. Hybridization of Labeled DNA Probes to Cytogenetic Specimens Hybridization of a cyotgenetic specimen with the probe requires heat-denaturation of the DNA probe in the hybridization buffer, and a pretreated and denatured slide specimen; together they are permitted to hybridize at 37◦ C. This protocol can be applied to cytogenetic specimens that are unstained or were previously G-banded (see Alternate Protocol 2). BASIC PROTOCOL 3 Materials DNA probe (see Basic Protocol 1 and Alternate Protocol 1) Pretreated and denatured cytogenetic slide specimen (see Basic Protocol 2 and Support Protocol 2) Rubber cement 75◦ C water bath or PCR machine 37◦ C dry oven Glass coverslips Hybridization box, slightly dampened (e.g., black plastic video tape box lined with slightly moistened gauze or paper towel) Hybridization container (i.e., any plastic container with lid or a black video cassette box) 1. Heat-denature the required amount of labeled DNA probe 5 min in a 75◦ C water bath or PCR machine. Preanneal 1 hr in a 37◦ C dry oven (see Critical Parameters). 2. Add the appropriate amount of DNA probe to the pretreated and denatured cytogenetic slide specimen using guide given in Table 22.4.11. Coverslip and ring the perimeter with rubber cement to seal in place. Table 22.4.11 Guide to Appropriate Probe Volume for a Given Coverslip Size Probe volume Coverslip size (mm) 10 µl 22 × 22 20 µl 22 × 30 30 µl 22 × 50 Table 22.4.12 Examples of Minimal Hybridization Times Required for Representative DNA Probes Minimal hybridization time (37◦ C) Probe type (300–500 bp) Centromere Probesa 4 hr a Subtelomere chromosome-specific probes 24 hr Chromosome paints 4 hr Translocation junction unique-sequence probes 24 hr Microdeletion unique-sequence probes 24 hr Probes detecting gene amplification 24 hr a Protein nucleic acid (PNA) probes, which are commercially available, require a minimum of 60 min for hybridization. Cell Biology of Chromosomes and Nuclei 22.4.17 Current Protocols in Cell Biology Supplement 23 3. Place slide in a slightly dampened hybridization box. 4. Transfer slide to a hybridization container and incubate overnight in a 37◦ C dry oven (see Table 22.4.12). 5. Proceed to post-hybridization washes and immunofluorescent detection of indirectly (see Basic Protocol 4) or directly labeled DNA probes (see Alternate Protocol 8) ALTERNATE PROTOCOL 5 Hybridization of PNA Probes to Cytogenetic Specimens PNA probes, as discussed below (see Critical Parameters), can be commercially obtained. Use of PNA probes requires minor modifications in the hybridization to both cytogenetic specimens and paraffin-embedded sections, as described below. Additional Materials (also see Basic Protocol 3) Labeled PNA probe (Applied Biosystems) Pretreated cytogenetic specimen, not denatured (Basic Protocol 2) 80◦ C oven, hot plate, or HYBrite (Vysis) 1. Add the required amount of labeled PNA probe to the pretreated, undenatured, cytogenetic specimen. 2. Coverslip and seal with rubber cement. Allow the rubber cement to set and dry. 3. Place the slide in an 80◦ C oven or HYBrite, or on an 80◦ C hot plate for 90 sec. 4. Remove the slide and place it into a hybridization box. Hybridize at least 1 hr at room temperature. 5. Proceed to post-hybridization washes for specimens hybridized with PNA probes (see Alternate Protocol 10). ALTERNATE PROTOCOL 6 Hybridization of Labeled DNA Probes to Sections from Paraffin-Embedded Material Unlike cytogenetic specimens, tissue sections from paraffin-embedded material require higher denaturation temperatures and longer denaturation times. Hybridization to these sections requires co-denaturation of the probe and tissue section simultaneously. All experiments using previously paraffin-embedded material and a DNA probe should include a minimal 24-hr hybridization. Additional Materials (also see Basic Protocol 3) Deparaffinized and enzyme-digested specimen (see Alternate Protocol 3) 90◦ C oven, hot plate, or HYBrite (Vysis) 1. Apply DNA probe to the deparaffinized and enzyme-digested specimen using the guide given in Table 22.4.11. Add a glass coverslip and apply rubber cement along the perimeter of the coverslip. Allow the rubber cement to dry. 2. Denature the probe and target DNA simultaneously by placing the slide in a 90◦ C oven or HYBrite, or on a 90◦ C hot plate for 12 min. Fluorescence In Situ Hybridization (FISH) 3. For slide denatured in an oven or on a hot plate: Transfer the slide to a hybridization container lined with a wet paper towel or damp gauze, and incubate overnight in a 37◦ C dry oven. 4. For slide denatured in a HYBrite: Program the unit to hold at 37◦ C. Incubate overnight. 22.4.18 Supplement 23 Current Protocols in Cell Biology 5. Proceed to post-hybridization washes and immunofluorescent detection of indirectly or directly labeled DNA probes (see Basic Protocol 4 or Alternate Protocols 8 or 9). Hybridization of PNA Probes to Sections from Paraffin-Embedded Material The following protocol describes hybridization of commercially available PNA probes to pretreated sections from paraffin-embedded samples. Unlike DNA probes, PNA probes require a minimum of 60 min for hybridization. ALTERNATE PROTOCOL 7 Additional Materials (also see Basic Protocol 3) PNA probes (Applied Biosystems) Deparaffinized and enzyme-digested specimen (see Alternate Protocol 4) 80◦ C oven, hot plate or HYBrite (Vysis) 1. Apply the appropriate amount of PNA probe (as suggested by the manufacturer) to the deparaffinized and enzyme-digested paraffin section using the guidelines provided in Table 22.4.11. Coverslip and seal with rubber cement. Allow the rubber cement to dry. 2. Place in an 80◦ C oven or HYBrite, or on an 80◦ C hot plate for 3 min. 3. Place in a hybridization container and allow to hybridize at least 1 hr at room temperature. 4. Proceed to post-hybridization wash for slides hybridized with PNA probes (see Alternate Protocol 10). POST-HYBRIDIZATION WASHES AND DETECTION Following hybridization, unbound probes, whether DNA or PNA, must be removed from the specimen. This is accomplished through washes of appropriate stringency, using formamide and SSC in varying amounts. After immunodetection, final washes contain detergents to remove unbound antibodies. The slides are counterstained and mounted in an antifade medium for visualization. Variations on washing procedures reflect the nature of the probe, whether directly or indirectly labeled. Post-Hybridization Washes and Immunofluorescent Detection of Indirectly Labeled DNA Probes This protocol describes post-hybridization washes and detection of indirectly labeled DNA probes. Following overnight hybridization, the coverslip is removed and the slide is immersed in a solution of formamide to remove any unbound probe. The slide is then washed in stringent SSC washes and blocked with a solution of BSA. Depending on the type of DNA probe used, amplification of the signal may be required. This is achieved by the addition of primary and secondary antibodies that may or may not be conjugated with a fluorochrome. Detergent washes are carried out after each antibody incubation; it is critical that the slide not be permitted to dry out at any point of the assay. The slide is then counterstained and ready for visualization. In multicolor FISH experiments, the user may have both indirectly and directly labeled probes on the same specimen. If this is the case, it is important to keep the ambient light dim to prevent quenching of the signal; Coplin jars with lids are useful for this purpose. If using multiple indirectly labeled probes, be sure that the primary and secondary antibodies are raised in different animals such that there is no cross-reaction. This protocol is applicable to cytogenetic slides or sections from paraffin-embedded samples; however, more stringent or additional washes may be required for paraffin experiments. BASIC PROTOCOL 4 Cell Biology of Chromosomes and Nuclei 22.4.19 Current Protocols in Cell Biology Supplement 23 Table 22.4.13 Coplin Jars Needed for Post-Hybridization Washes and Immunofluorescent Detection of Indirectly Labeled DNA Probesa No. Coplin jars Contents Temperature 3 50% formamide in 2× SSC 45◦ C 3 1× SSC 45◦ C 9 0.1% Tween 20 in 4× SSC 45◦ C a See Basic Protocol 4. Materials Hybridized slides in a hybridization box (see Basic Protocol 3 and Alternate Protocol 6) 50% formamide in 2× SSC, 45◦ C (prepare fresh) 1× SCC, 45◦ C (UNIT 18.6) Blocking solution: 1% BSA (w/v)/0.1% (v/v) Tween 20 in 4× SSC (store indefinitely at −20◦ C or up to several months at 4◦ C) Primary, secondary, and tertiary antibodies (see recipe for antibodies) 0.1% (v/v) Tween 20 in 4× SSC, 45◦ C DAPI in antifade (see recipe) Clear nail polish Coplin jars 22 × 50–mm glass coverslips 37◦ C oven Fluorescent microscope and appropriate filter sets (UNIT 4.2) CAUTION: Formamide is a carcinogen and should be handled with care. Discard according to biohazard rules of the institution. NOTE: A list of all Coplin jars used in this protocol is given in Table 22.4.13. Remove coverslip 1. After 24 hr, remove hybridized slide from the hybridization box. 2. Carefully peel the rubber cement from the hybridized slide and immerse in a Coplin jar containing 50% formamide in 2× SSC, 45◦ C. 3. Allow the coverslip to fall off and let stand 5 min in solution with gentle agitation. Wash and block 4. Remove slide and transfer to second Coplin jar containing formamide/SSC, 45◦ C, for 5 min. Agitate gently. Repeat with a third Coplin jar containing formamide/SSC, 45◦ C. 5. Wash the slide for 5 min each in three consecutive Coplin jars containing 1× SSC, 45◦ C, agitating gently between washes. 6. Drain excess solution but do not allow the slides to dry. Add 80 µl blocking solution, coverslip with 22 × 50–mm glass, and place back in hybridization box. Incubate 40 min at 37◦ C. Fluorescence In Situ Hybridization (FISH) Label with primary antibody 7. Remove coverslip and add 80 µl primary antibody. Coverslip and place back into the hybridization box. Incubate 40 min at 37◦ C. 22.4.20 Supplement 23 Current Protocols in Cell Biology Table 22.4.14 Common Primary and Secondary Antibody Systems and Final Concentrationsa Hapten Primary antibody Secondary antibody Tertiary antibody Color Probe Type Biotin (no signal amplification) 5 µg/ml FITC-avidin — — green Centromere chromosome paints Biotin 5 µg/ml FITC-avidin 5 µg/ml biotinylated 5 µg/ml FITC-avidin Green goat anti-avidin Locus-specific cDNAs Biotin 5 µg/ml rhodamine-avidin 5 µg/ml biotinylated 5 µg/ml goat anti-avidin rhodamine-avidin Red Locus-specific cDNAs Digoxigenin (no 2 µg/ml rhodamine signal amplification) anti-Dig — — Red Centromere chromosome paints Digoxigenin (no 2 µg/ml FITC signal amplification) anti-Dig — — Green Centromere chromosome paints Digoxigenin 0.5 µg/ml mouse anti-digoxigenin 2 µg/ml digoxigenin 2 µg/ml rhodamine anti-mouse anti-Dig Red Locus-specific cDNAs Digoxigenin 0.5 µg/ml mouse anti-digoxigenin 2 µg/ml digoxigenin 2 µg/ml FITC anti-mouse anti-Dig Green Locus-specific cDNAs a Shown in this table are the most common antibody systems for the detection of biotinylated and digoxigenin (Dig)-labeled DNA. Final concentrations for antibodies are also stated but may require adjustment. A variety of antibody systems are available from Molecular Probes and Roche Diagnostics. If a two–color FISH approach is being used, be sure that respective primary, secondary, and tertiary antibodies do not cross-react. This may require sequential hapten detection rather than concomitant hapten detection. The scheme above uses antibodies raised against mouse and goat, thus no cross-reaction will occur. Antibodies raised in rabbits will also offer more variety in hapten detection. This antibody may or may not have a fluorescent moiety conjugated to it, depending on the nature of the probe (see Critical Parameters). Common primary antibody and secondary antibody systems are outlined in Table 22.4.14. 8. Remove coverslip and wash slide for 5 min each in three consecutive Coplin jars containing 0.1% Tween 20/4× SSC, 45◦ C, with gentle agitation. If proceeding with signal amplification (see Table 22.4.15) continue to step 9. Otherwise, proceed to step 14. 9. Drain excess wash solution, but do not allow slide to dry. Add 80 µl blocking solution, coverslip, and place back into the hybridization box. Incubate 10 min at 37◦ C. Label with secondary antibody 10. Remove coverslip and add 80 µl secondary antibody (Table 22.4.14). Coverslip and place back into the hybridization box. Incubate 30 min at 37◦ C. 11. Remove coverslip and wash slide 5 min each in three consecutive Coplin jars containing 0.1% Tween 20 in 4× SSC, 45◦ C, with gentle agitation. Label with tertiary antibody 12. Remove coverslip and add 80 µl tertiary antibody (conjugated to fluorochrome). Coverslip and place back into the hybridization box. Incubate 30 min at 37◦ C. 13. Remove coverslip and wash slide 5 min each in three consecutive Coplin jars containing 0.1% Tween 20 in 4× SSC, 45◦ C, with gentle agitation. Counterstain and seal 14. Drain excess wash solution, but do not allow the slides to dry. Add 40 µl DAPI in antifade counterstain. Coverslip and seal with clear nail polish. The slides are now ready for visualization. When not in use, store slides up to several weeks or months at −20◦ C, depending on the frequency of visualization. 15. Examine using a fluorescent microscope and appropriate filter sets. Cell Biology of Chromosomes and Nuclei 22.4.21 Current Protocols in Cell Biology Supplement 23 Table 22.4.15 Interpretation of Parallel Positive Control Experiments Problem Slide hybridized with new probe Slide hybridized with good known Analysis probe Background Present Not present New probe may still contain unincorporated dNTPs or many small labeled fragments are present and did not hybridize. Slide quality is not the problem. Washing conditions also do not appear to be the cause. Background Present Present Problems likely related to washing conditions or slide quality rather than issues with the probe. Weak Signal (indirect-labeled probe) Yes No If using antibody detection system for indirectly labeled probes, this scenario indicates that the antibodies are in good working order. Look to problems with incorporation of the hapten into the DNA. Weak signal (direct-labeled probe) Yes No This indicates a problem with the labeling of the probe. Check that all enzymes and fluorochromes are within their shelf life. This also indicates that washing conditions are correct. Weak signal (indirect labeled probe) Yes Yes This indicates a problem with the antibody detection system. Ensure that the concentrations are correct and that the antibodies are fresh. It may also indicate a problem with hybridization, insufficient denaturation of the target DNA on the slide, or overall quality of the DNA specimen. Weak signal (direct-labeled probe) Yes Yes The problem may be in the post-hybridization washes: too stringent or temperatures too high. Poor chromosome morphology with weak signal Yes Yes Over denaturation if the chromosomes appear puffy. Good chromosome morphology but no signal Yes Yes Slides are likely too old and resistant to denaturation. Change to a more fresh preparation. Otherwise the experiment can be repeated but the denaturation of the slide should be increased. ALTERNATE PROTOCOL 8 Post-Hybridization Washes and Detection of Directly Labeled DNA Probes The protocol below outlines steps for washing slides hybridized with directly labeled probes. If using commercially produced probes, follow the manufacturer’s instructions. This protocol is applicable to hybridized cytogenetic slides or paraffin sections. See Basic Protocol 4 for materials. NOTE: A list of all Coplin jars used in this protocol is given in Table 22.4.16. Fluorescence In Situ Hybridization (FISH) 1. After 24 hr hybridization, carefully peel off rubber cement from slide and immerse in a Coplin jar containing 50% formamide in 2× SSC, 45◦ C. Allow the coverslip to fall off and let stand 5 min. 2. Remove slide and transfer to a fresh Coplin jar containing formamide/SSC, 45◦ C. Incubate 5 min. Repeat with a third Coplin jar containing formamide/SSC, 45◦ C. 22.4.22 Supplement 23 Current Protocols in Cell Biology Table 22.4.16 Coplin Jars Needed for Post-Hybridization Washes and Detection of Directly Labeled DNA Probesa No. Coplin jars Contents Temperature 3 50% formamide in 2× SSC 45◦ C 3 1× SSC 45◦ C 3 0.1% Tween in 4× SSC 45◦ C a See Alternate Protocol 8. Table 22.4.17 Coplin Jars Needed for Rapid Wash of Directly Labeled Probesa No. Coplin jars Contents Temperature 1 0.3% (v/v) NP-40 in 2× SSC 72◦ C 1 0.3% (v/v) NP-40 in 2× SSC Room temperature 1 2× SSC Room temperature a See Alternate Protocol 9. 3. Wash slide 5 min each in three consecutive Coplin jars containing 1× SSC, 45◦ C. 4. Wash slide 5 min each in three consecutive Coplin jars containing 0.1% Tween 20 in 4× SSC, 45◦ C, with gentle agitation. 5. Drain excess wash solution, but do not allow the slide to dry. Add 40 µl DAPI in antifade counterstain. Coverslip with 22 × 50–mm glass and seal with clear nail polish. Store slide up to several months at −20◦ C. Fluorescence fading will depend upon the frequency of viewing. 6. Examine using a fluorescent microscope equipped with the appropriate filters. Rapid Wash of Directly Labeled Probes This protocol describes the use of high temperatures and stringent SSC washes for the removal of unbound probe from target DNA. This protocol is generally effective for directly labeled DNA, particularly from commercial sources. It is important to consider the differing stringency requirements of each probe being used when planning this protocol since excessive temperature can strip bound probe from the target. This protocol is applicable to cytogenetic slides or paraffin sections. ALTERNATE PROTOCOL 9 Additional Materials (also see Basic Protocol 4) 2× SSC (UNIT 18.6) 0.3% (v/v) NP-40 in 2× SSC, room temperature and 72◦ C NOTE: A list of all Coplin jars used in this protocol is given in Table 22.4.17. 1. Carefully remove rubber cement from the hybridized slide. Place the slide and coverslip in a Coplin jar containing 2× SSC to gently remove coverslip. 2. Once the coverslip has fallen off, incubate the slide 1 to 2 min in a Coplin jar containing 0.3% NP-40 in 2× SSC, 72◦ C. 3. Remove slide and wash 1 min in a Coplin jar containing 0.3% NP-40/2× SSC, room temperature. 4. Drain solution from slide and add DAPI in antifade counterstain. Add a 22 × 50–mm glass coverslip and seal with clear nail polish. 5. Examine the slide with a fluorescent microscope equipped with the appropriate filters. Cell Biology of Chromosomes and Nuclei 22.4.23 Current Protocols in Cell Biology Supplement 23 ALTERNATE PROTOCOL 10 Post-Hybridization Washes for Specimens Hybridized with PNA Probes This protocol describes a procedure for removing unbound PNA probe from cytogenetic specimens or from paraffin sections. Materials Slides hybridized with PNA probes (see Alternate Protocol 5 or 7) 70% (v/v) formamide/10 mM Tris·Cl (pH 7.0 to 7.5)/0.1% (w/v) BSA (see recipe) 0.1 M Tris·Cl (pH 7.0 to 7.5)/0.15 M NaCl/0.08% (v/v) Tween 20 (store up to several weeks at room temperature) 70%, 90%, and 100% ethanol DAPI in antifade solution (see recipe) Fluorescent microscope and appropriate filters NOTE: A list of all Coplin jars used in this protocol is given in Table 22.4.18. 1. Remove coverslip from slide hybridized with PNA probes and wash 15 min in a Coplin jar containing 70% formamide/10 mM Tris·Cl (pH 7.0 to 7.5)/0.1% BSA. Repeat once. 2. Wash 5 min each in three consecutive Coplin jars containing 0.1 M Tris·Cl (pH 7.0 to 7.5)/0.15 M NaCl/0.08% Tween 20. 3. Dehydrate slide by incubating 5 min each in Coplin jars containing 70%, 90%, and 100% ethanol. Air dry (5 to 10 min). 4. Apply DAPI in antifade and coverslip. 5. Examine the slide using a fluorescent microscope and appropriate filters. 6. Store slides up to several months at −20◦ C INTERPRETATION OF FISH FINDINGS This section discusses interpretation of FISH experiments and relevant troubleshooting measures. Each laboratory may adopt a different means of assessing positive and negative results as is applicable to the experiment and the question being asked. The guidelines below can generally be applied to both interphase and metaphase analysis. Evaluating FISH FISH encompasses four main components: the slide and its preparation (see Target Slide Preparation), the DNA probe and its preparation (see Labeling DNA Probes for FISH), Table 22.4.18 Coplin Jars Needed for Post-Hybridization Washes for Specimens Hybridized with PNA Probesa No. Coplin jars Fluorescence In Situ Hybridization (FISH) Contents Temperature 2 70% formamide/10 mM Tris·Cl (pH 7.0–7.5)/0.1% BSA Room temperature 3 0.1 M Tris·Cl (pH 7.0–7.5)/0.15 M NaCl/0.08% Tween 20 Room temperature 1 70% ethanol Room temperature 1 90% ethanol Room temperature 1 100% ethanol Room temperature a See Alternate Protocol 10. 22.4.24 Supplement 23 Current Protocols in Cell Biology denaturation and hybridization of the probe to the slide (see Hybridization), and finally, post-hybridization washes and detection (see Post-Hybridization Washes and Detection). Each of these components is, in itself, multistepped. The discussion below outlines many of the problems encountered in FISH experiments, including factors influencing signal strength, background, and preserving optimal chromosome morphology. Other sources for FISH optimization and parameters can be found in Beatty et al. (2002), Henegariu et al. (2001), Schwarzacher and Heslop-Harrison (2000), and van de Rijke et al. (1996). Signal strength Slide age. In the author’s experience, optimal results have been obtained from slides not older than 2 months. As the slides age further (3 to 6 months), they become harder to denature. Conversely, very old preparations (>1 year) often have partially degraded DNA that may reduce or preclude effective FISH experiments. In such situations, preparations tend to become very sensitive to denaturation. Previously G-banded slides (UNIT 22.3) have an even shorter life span and should be processed within 2 weeks (refer to Table 22.4.5). Cytoplasmic debris. The presence of cytoplasm on the slide may inhibit binding and contribute to overall background. A more aggressive protease pretreatment may be required to reduce cytoplasmic protreinacious noise. This can be accomplished by increasing the time of digestion or the amount of protease added (maintain the same digestion time). One must also consider whether the slide is made from a dropped suspension or FISHed to a slide made from an in situ culture, since the latter slides tend to possess more cytoplasmic and cellular debris. A high background obscures true signals. If there is minimal background but the signal is weak in different parts of the slide, then there may be a gradient or patchiness of cellular protein in areas of the slide with a higher density of fixed cells. Excessive slide pretreatment. Excessive enzymatic treatment may damage the target DNA, making it less efficient for hybridization with the probe. This is usually indicated by weak uptake of the counterstain and the presence of bright centromeres. Denaturation time. As the slide ages (see above), the chromosomes become harder to separate into single strands. Slides used within 1 to 2 weeks of preparation should be denatured for 1.5 to 2 min. Slides that are older may require times that range from 2 to 3 min. Under-denaturaton results in insufficient strand separation of the target DNA, decreasing the effective hybridization efficiency. Over-denaturation of the target DNA causes DNA damage and reduces the amount of target DNA that is able to hybridize with the probe; it also results in reduced hybridization efficiency. Over-denaturation usually also results in poor uptake and banding using the DNA counterstain and very bright centromeres. Sealing of coverslip. It is critical that the coverslip be adequately sealed with rubber cement during incubations to prevent any moisture from entering the hybridized area, thus diluting the probe. Do not use contact cement or other ultra-adhesive glues. Proper temperature for washes and incubations. Check that the temperature in the oven, hot plate unit, or water bath is correct for hybridization and other incubations. High temperatures during post-hybridization washes may remove bound probe with weak sequence homology (see Critical Parameters), thus decreasing the amount of DNA available for signal detection or antibody binding. Avoid taking shortcuts during incubation times with blocking reagents or antibody detection reagents. Also be sure that the reagents do not dry up on the slide; this makes washing more difficult. Detergent type and concentration. Increasing the detergent content or choice of detergent may influence signal strength. Tween 20 is generally less harsh than NP-40. Increasing Cell Biology of Chromosomes and Nuclei 22.4.25 Current Protocols in Cell Biology Supplement 23 the detergent concentration in combination with increasing temperature will remove more probe and/or bound antibodies. Be sure that the concentration and temperatures are correct. SSC concentration. Decreasing SSC concentration increases stringency. Thus 0.1× SSC destabilizes double-stranded DNA more readily than 2× SSC. Double-stranded DNA stability is also affected by the extent of sequence homology between the probe and target DNAs. Ensure that the proper SSC concentrations are used during washes. Proper filter sets. For fluorescence, make sure that the proper filter sets are used for image acquisition. The use of filters with incorrect spectral characteristics for the fluorochrome being used can severely impair the ability to detect the correct signals or they may increase the amount of autofluorescence. Also refer to UNIT 4.2 and APPENDIX 1E. Amplification of signal. When using an indirectly labeled DNA probe, signal amplification may be required. If the signal is amplified but still weak, check the concentrations and shelf lives of antibodies used. If the antibodies are in good order, relabel the probe and check the labeling efficiency (see Critical Parameters). For directly labeled probes, consider the labeling efficiency of the probe. Probe characteristics In-house probes. If probes are labeled in-house (see Labeling DNA Probes for FISH), strict controls must be undertaken to ensure that proper hapten or fluorochrome incorporation has been obtained to produce a high-quality DNA probe. Haptens may be assessed using the dot blot method of incorporation. Directly labeled probes can be assessed by removing a small aliquot of probe, placing it on a slide, and viewing it by fluorescence microscopy. Cot-1 suppression. Excessive Cot-1 suppression may be the cause of reduced signal. Smaller DNA probes or cDNAs contain fewer repeat elements compared to larger inserts. Normally, 5 to 10 µg Cot-1 is sufficient per 100 ng labeled DNA; adjust as required. Commercial probes. Usually commercial suppliers properly process the product with the necessary quality controls. Check that the probes were properly stored and used before the expiration date. For some experiments it is possible to optimize procedures using concentrations of commercial probes at levels slightly below those recommended by the supplier. This can prove very cost effective if a specific commercial probe is going to be used routinely. Probe concentration. The probe is usually used in excess of the target DNA; however, make sure that sufficient probe volume has been added to adequately cover the area of interest on the slide (see Table 22.4.11). Preannealing. Preannealing at 37◦ C is usually carried out for 1 hr following denaturation. This step is performed to permit the annealing of repetitive sequences in the DNA probe with unlabeled Cot-1 DNA to prevent cross-hybridization. If there are few repetitive elements in the probe, or the probe is a cDNA or small insert, this step may be omitted. The probe is simply denatured and applied to the denatured slide. Background Fluorescence In Situ Hybridization (FISH) Cytoplasmic debris. This is the most common cause of background. Increase the incubation time during protein digestion or maintain the same time but change the concentration of the enzyme. 22.4.26 Supplement 23 Current Protocols in Cell Biology Bacterial/yeast contamination. Microorganisms that contaminate cultures used to make the slide, or reagents used to prepare the slide or for hybridization are deposited onto the slide. If the contamination is minor, then it is only necessary to analyze the slide in areas that contain fewer deposits of fungi or bacteria. If the contamination is heavy, the slide or reagent must be prepared again. Coverslips. Coverslips should be clean and dust free. If cells are to be grown in situ, use only glass coverslips as plastic coverslips autofluoresce. Residual oils. Slides that have been previously visualized using immersion oil (e.g., Gbanded slides) should be cleaned with xylene. Residual oils will prevent hybridization and cause background problems. Dust and other particles. Particles may be deposited on the slide during various transfers into solutions. Be sure that solutions are well mixed and filtered as needed. Some solutions may form precipitates that may bind to the slide. In-house probes. Ensure that nonincorporated conjugated nucleotides are removed from the final probe preparation. Commercial probes. These probes are usually not the cause of background, especially when the same probe on another slide preparation has not produced background; however, note the lot number for future reference. It is also very helpful to introduce a new batch of a commercial probe into routine use before the previous lot is exhausted in case the supplier has had production difficulties Unlabeled DNA: carrier and Cot-1. An excess of unlabeled DNA is added to the probe mixture. Although it is unlabeled, it can contribute to background by trapping antibodies. To reduce this problem, ensure that the resuspended probe pellet is fully dissolved in sufficient hybridization buffer. If the pellet does not completely dissolve, add more buffer. This will not significantly alter the reaction. Denaturation time. The probe is usually heat denatured a minimum of 5 min. This should be sufficient time to denature the probe and dissolve any remaining DNA. The denaturation time may be increased to 10 min without any damage to the DNA probe. Post-hybridization washes. Make sure that the correct temperature has been maintained for washes and incubations. Agitation during the washes can help to remove unbound probe and antibodies. Increasing the stringency of the washes by either increasing the temperature or altering the amount of SSC in the washes can also help considerably if background is encountered. Avoid drying the slide with any residual blocking or detection reagents. Detergent concentration. Increasing the detergent concentration in post-hybridization washes in combination with increasing temperature removes more probe and/or bound antibodies. Be sure that the concentration and temperatures are correct. Fading signals In-house probes. Ensure that stocks of directly conjugated dNTPs are properly stored in the dark and used well before the expiration date. Unlike indirectly labeled probes, directly labeled DNAs have a much shorter shelf life. Commercial probes. Although these probes are supposed to be quality controlled, these too are subject to the same concerns as in-house probes (see above). During denaturation Cell Biology of Chromosomes and Nuclei 22.4.27 Current Protocols in Cell Biology Supplement 23 and hybridization, keep lights dimmed and hybridize in a light-proof container for directly labeled probes. Directly-labeled DNA probes. When directly labeled probes are used, slides should be washed with the lights dimmed to prevent unnecessary exposure to extraneous light. Mounting medium. Check that the DAPI/antifade mounting solution is used before its expiration date. Normally, the medium is clear with a slight pink tinge; medium that is degrading will turn increasingly amber. Expired antifade medium causes rapid signal degradation and displays a red glow when viewed under the microscope. Image acquisition. Extended exposure of the hybridized slide to UV sources leads to signal quenching. When not in use, store slides at −20◦ C. During visualization, keep the light source off when not in use. Poor chromosome morphology Slide age. See Signal Strength (above) for information concerning this parameter. Cytoplasmic debris. See Signal Strength (above) for information concerning this parameter. Excessive slide pretreatment. Excessive enzymatic treatment may damage the target DNA, making it less efficient for hybridization with the probe Previously G-banded slides. G-banding (UNIT 22.3) causes DNA to become sensitive to subsequent denaturation. For this reason, denaturation times are greatly reduced. This also has an effect on the quality of hybridization and signal strength. Over-denaturation. The cause of poor chromosome morphology is usually overdenaturation, which causes the DNA to be destroyed. This can be sample specific and/or slide-age related. Denaturation temperature. Check that the temperature for denaturation is accurate. The final internal temperature of the Coplin jar should be 72◦ C. Add 1◦ C for each slide that is denatured in the jar. Avoid denaturing more than five slides in one jar at one time. Note that plastic Coplin jars have a greater differential temperature between bath and internal temperatures as compared to glass. Probe cross-hybridization Clone identity. When using in-house FISH probes for research applications, ensure that the clone identity and the insert are correct for the experiment. Some genes have highly homologous sequences elsewhere in the genome and may cross-hybridize to other family members. DNA contamination. Consider whether stock DNA was contaminated with DNA from another clone. Increase the hybridization temperature if contamination is suspected. Typically, the hybridization temperature is 37◦ C. If cross-hybridization occurs, try increasing the hybridization temperature to 42◦ C. Fluorescence In Situ Hybridization (FISH) Stringency of washes. Some probes may cross-hybridize, particularly centromere probes. Usually cross-hybridization signals are significantly weaker than true signals. More stringent washes can be undertaken by decreasing the SSC concentration, increasing wash temperature, or adding additional washes. 22.4.28 Supplement 23 Current Protocols in Cell Biology Evaluation of a New DNA Probe The most critical factor in optimizing a new FISH probe which was labeled in-house is first determining whether the probe of interest maps to the correct location. Many vectors (e.g., plasmids, cosmids, BACs) have traveled from laboratory to laboratory and have changed hands many times. These DNAs, as well as cloned DNA obtained from reputable repositories, can be mislabeled or misidentified. After a cloned DNA has been extracted and labeled, the first FISH experiment should be carried out on normal lymphocyte spreads and should address the following questions. Is a signal visible on a pair of chromosomes? If there is a weak signal, refer to the section above. Does the DNA probe map to the right chromosomal location? Does it map to only one location? It helps to have someone who can identify the chromosomes by DAPI banding. Most people are not as proficient as trained cytogeneticists, thus it may be useful to carry out a two-color experiment using a commercially available centromere probe for the chromosome of interest. Refer to UNIT 22.3 for general conventions for identification of chromosomes according to ISCN nomenclature. Some DNA sequences/genes may belong to families with similar sequences. It is possible that hybridization to these similar regions may occur. Usually the true signals are stronger than the cross-hybridized ones. If this is the case, wash conditions can be adjusted or another clone should be obtained. Does a positive control probe work properly? It is useful to perform parallel experiments with a probe known to give good signals in the same experiment. This will help to identify any problems that are not related to the newly labeled probe (see Table 22.4.15 for interpretation of such control experiments). For example, in panel iii of Figure 22.4.4A, two mouse BAC probes were labeled (one green and the other red) and assessed. A separate slide with a previously labeled BAC known to give good signal and no background was prepared as a control at the same time (not shown). The newly labeled BAC probes shown in panel iii of Figure 22.4.4A mapped to the proper location with weak signal intensity and high background. Analysis of this experiment is as follows. The BAC probes map to the correct location. Because the signal intensity was weak consider (1) starting DNA quality and quantity, (2) labeling efficiency, (3) fidelity of DNA polymerase I and DNase I, (4) labeling time and temperature, (5) effective removal of nonincorporated nucleotides, and (6) insufficient Cot-1 suppression. Because the control probe displayed no background problems, one can assume that the hybridization and wash conditions were sufficient. Evaluation of Hybridization Efficiency The minimum number of cells or metaphase spreads required to obtain a given result reflects the clinical context of the finding, the limitations of the patient material available for study, and the question being asked. With tissues or cells that are hard to obtain, a single abnormal metaphase may be significant. For example, in some situations, limited FISH data may be supported by results obtained using PCR and/or Southern blot analysis. Prior to enumerating or analyzing FISH results on a test sample, it is important to carefully assess the overall quality, uniformity, and effectiveness of hybridization as discussed Cell Biology of Chromosomes and Nuclei 22.4.29 Current Protocols in Cell Biology Supplement 23 Figure 22.4.4 Legend at right. Fluorescence In Situ Hybridization (FISH) 22.4.30 Supplement 23 Current Protocols in Cell Biology above. Each hybridized slide should be evaluated for the specificity of the hybridization, the probe signal intensity, and the signal-to-background noise, to determine if the hybridization was optimum for the given analyses. There should be minimal background or nuclear fluorescent noise. At least 85% of all nuclei in the target area should be easily innumerable. For some applications (such as the detection of mosaicism or minimal residual disease), more rigorous analytical sensitivities and hybridization efficiencies are required. 1. Perform the FISH protocol appropriate for the type of patient and control slides in the experiment. 2. Prior to determining the hybridization efficiency, quickly scan the whole slide, noting the general signal-to-noise levels in different regions of the slide and any areas with high background or unusually weak signals. It may be useful to mark the underside surface of the slide in these areas with a diamond pen. If the signal intensity far exceeds background levels, proceed with estimating the hybridization efficiency. 3. Pick several representative areas of the slide and score at least 200 nuclei from the areas selected, following the selection criteria described below in Selecting Cells for FISH Microscopy. Keep a running log of the number of cells scored and the observed signal counts for the patient and control slides. 4. For all slides, add up the number of cells with no signal. In general, hybridization is considered to be adequate if >85% of the cells scored have one or more signals. Lower hybridization efficiencies may be encountered with smaller probes (generated Figure 22.4.4 (at Left) Composite figure illustrating FISH hybridization. (A) Assessment of a DNA probe. (i) Cross-hybridization of probe (arrowhead) to chromosome 1. True signals are located on chromosome 22. (ii) Changes in stringency washes and post-hybridization removes cross-hybridization. (iii) High background associated with a insufficient repetitive sequence (Cot1) suppression in this mouse metaphase spread. Weak signals (arrows) indicate poor labeling efficiency. (B) Interphase FISH analysis. (i) A cytogenetic specimen from a short-term ovarian primary culture hybridized with PNA probes for centromeres 7 (green) and 8 (red). When scoring interphase nuclei, it is especially important to focus through the cells since signals may be present at different z planes. (ii) Schematic indicating the nuclei that are acceptable for scoring: A, cell acceptable for scoring; B, cell is likely acceptable for scoring, but requires careful attention; ?, cell has qualities that make it questionable for scoring; X, cell is not acceptable for scoring. (iii) Interphase analysis using PNA probes specific for centromeres 7(green) and 8 (red) on a paraffin section from a prostate carcinoma. Arrowheads indicate cells containing changes in ploidy. (C) Analysis of translocation probes. (i) and (ii) Results of consecutive hybridization of a G-banded metaphase spread with the Vysis BCR/ABL translocation probe. Two fusion signals (yellow) are produced from the hybridization of BCR/ABL fusion on the Philadelphia chromosomes (Ph) on chromosome 22 and the reciprocal ABL/BCR on the derivative chromosome 9. Separate green and red signals from the normal chromosomes 9 and 22 are also seen. (iii) Interphase pattern from this specimen. (D) Example of a chromosomal inversion on chromosome 11. An inversion was identified involving the terminal portion of chromosome 11 by gross cytogenetic analysis. Clones 200 kb apart and in the 11p15.5 region, containing the IGF2 gene (green) and H19 (red) were hybridized to the patient specimen. The normal chromosome 11 shows the red and green signal hybridizing on top of each other at 11p15.5. The inverted 11 shows the clear spit of signal along the abnormal chromosome 11 indicating the breakpoint lies within the 200 kb between IGF2 and H19. (E) Gene amplification of MYCN in neuroblastoma specimens. (i) Double minute chromosomes (dmns) containing hundreds of copies of the MYCN gene. This is in contrast to amplification of MYCN in (ii) as a large block of signal called a homogeneously staining region (HSR). Interphase nuclei nearby show the typical hybridization pattern for an HSR. (F) Use of subtelomeric and pan-centromeric PNA probes. (i) Hybridization of a prostate cell line with PNA subtelomeric probes. (ii) Hybridization of another prostate cell line with subtelomeric and pan-centromeric PNA probes. Loss of telomeric sequences are indicated by the arrow while the presence of multicentric chromosomes are indicated by arrow heads. This black and white facsimile of the figure is intended only as a placeholder; for full-color version of figure, see color plates. Cell Biology of Chromosomes and Nuclei 22.4.31 Current Protocols in Cell Biology Supplement 23 for example in a research laboratory). Extreme caution must be exercised when using probes with lower hybridization efficiencies or elevated background signal to provide clinical information. Selecting Cells for FISH Microscopy Generally, look at all areas of the slide and analyze regions with uniformity in signal strength. Compare the intensity of the background signals to the intensity of the signals in the nuclei or metaphases of interest. The FISH signal intensity should consistently be greater than the background intensity in the regions of the slide chosen for analysis. If the background signals are equivalent to signals in the nuclei, then the counts will be skewed and the results biased. The following provides a guide to selecting targets for analysis. 1. Ideally analyze cells from all areas of the slide. Systematically select representative areas from different regions of the slide. Any regions that have unacceptable background or weak signals identified in prescreening evaluations should be excluded. 2. Select nuclei or metaphase cells that do not touch or overlap (Fig. 22.4.4B). 3. Select intact nuclei that have smooth well-rounded borders. Partially ruptured nuclear membranes may have lost informative chromatin. Similarly select metaphase spreads that have no evidence of preparation artifacts or breakage. 4. Select cells that are not surrounded by cytoplasmic material and that are without evidence of potential drying artifacts such as rings or clumped cells. 5. Do not evaluate interphase nuclei with signals located on the extreme periphery of the nucleus. 6. Do not score regions of the slide containing nuclei that have no signals. Absence of signals may represent uneven or patchy hybridization, resulting in some areas of the slide having very weak or absent signals. Analysis of Interphase Nuclei Interpretation of interphase FISH is very much dependent on statistical analyses and has inherent technical challenges. For instance, the presence of signal is dependent upon the probe and its fluorescent label successfully entering the cell and hybridizing to the target DNA. Detection of the correct number of signals can be complicated by signals overlapping or splitting. Any background hybridization whatsoever leads to major complications in interpretation. It is not uncommon to find monosomy or trisomy in nuclei that reflect technical artifact or false positive background signals. Therefore, the accuracy of interphase FISH analysis is dependent upon recognizing these technical issues, correcting for them, and standardizing the scoring criteria accordingly. Interphase analysis is typically used for enumeration of chromosomes using centromere probes, detection of gene amplification or deletion (see below), and detection of the presence of translocations, so that many cells should be scored. Metaphase FISH analysis, although more informative, is more difficult, especially when there are few metaphase spreads present. 1. Select nuclei in which signals generally have the same intensity. 2. Focus up and down in the z axis to accommodate spatial configurations of probe signals within the nucleus (Fig. 22.4.4B). Fluorescence In Situ Hybridization (FISH) 3. Signals that are more intense in some nuclei than the specific signal indicate the presence of regional background. Care must be taken when analyzing any sample with this type of background noise. 22.4.32 Supplement 23 Current Protocols in Cell Biology Figure 22.4.5 Schematic representation of scoring criteria. Shown in the illustration are typical hybridization signal configurations. These images pertain to a single probe, such as a centromere- or locus-specific probe. Signals must be more than one signal width apart to be considered one signal. Signals joined by a string of hybridization are also considered as one signal. 4. Do not count lower level nonspecific hybridization signals. These signals can usually be recognized by their lower intensity and different shape. 5. Some nuclei will have passed through the S phase of the cell cycle and may be present as G2-paired signals (i.e., two smaller signals in very close proximity). These paired signals represent a single chromosome already divided into chromatids and should be counted as one signal (Fig. 22.4.5). 6. Count two signals connected by a strand of fluorescence as one signal. Sometimes centromere or long genomic probes will generate signals that are not spherical. Typically, FISH signals appear as separate fluorescent dots on each chromatid of a metaphase chromosome when the target size is 100 to 250 kb. Similarly, in interphase nuclei, such probes will also generate discrete easy-to-interpret signals. Larger probes can appear as fused signals straddling both chromatids, and in interphase nuclei, these probes can generate signals that present more diffuse or dispersed hybridization spots in the chromatin of interphase nuclei. Knowledge of the probe size and anticipated configuration in both metaphase spreads, and interphase nuclei is essential. As long as the signal is continuous it should be scored as one signal. 7. Count only nuclei in which a definite enumeration can be made. Do not analyze or enumerate inconclusive cells. 8. Use two people to score 200 consecutive nuclei from each sample such that each person scoring will analyze ∼100 nuclei from a given sample. The slides should be coded and scored independently by two analysts. Any discrepancies may mean the established scoring criteria for the FISH assay are not being adhered to rigidly. Cell Biology of Chromosomes and Nuclei 22.4.33 Current Protocols in Cell Biology Supplement 23 Analytical sensitivity of interphase FISH assays Once a particular probe set is made available as a routine FISH test for a clinical service, it is important that the laboratory perform assay validation and create a database establishing reportable range and general laboratory experience with each probe. The analytical sensitivity assay measures the success of a given FISH test in a particular laboratory environment and on a given tissue type. Since there are known differences in the cell populations and tissue types, it is important to use the appropriate positive control tissue for the assessment of analytical sensitivity and for the establishment of the database. Analytical sensitivity analyses are performed by scoring 200 interphase nuclei representing at least five normal, preferably male, individuals. (Pooling samples on one slide is acceptable.) The nuclei are scored for the percentage of nuclei exhibiting the appropriate number of distinct signals. For constitutional studies using FISH, the recommended analytical sensitivity for probes intended for nonmosaic detection is 90%, while for probes intended to detect mosaicism, it is 95%. Similarly, for detection of minimal residual disease, sensitivities ≥95% are helpful. Databases for each probe should be established so that it will then possible to determine the mean and standard deviation of results from a series of normal samples processed and analyzed in the same manner as clinical samples. False positive rates can then be calculated and used for final scoring reports. More discussion on this issue is available from the following sources: VYSIS guidelines for single (http://www.vysis.com/tech sup fishproto quality single.asp) and dual probes (http://www.vysis.com/tech sup fishproto quality dual.asp), and scoring criteria for preimplantation genetic diagnosis of numerical abnormalities for chromosomes X, Y, 13, 16, 18, and 21 (Munne et al., 1998). Statistical considerations concerning interphase FISH analysis of paraffin sections Due to truncation of the nuclei during sectioning, loss of signal from areas of the nucleus excluded from the target slide will be encountered when enumerating signals after FISH has been performed on paraffin sections. The criteria for determining the significance of loss or gain of signals in interphase nuclei will depend on a number of parameters (e.g., nuclear diameter, age of patient, type of tissue). Readers are referred to some of the scientific literature where suggested cutoff values are adopted from the available literature (Qian et al., 1996). In the authors’ experience with FISH analysis of prostate cancer, chromosomal gains can been identified when more than ∼10% of the nuclei exhibit more than two signals. Panel iii of Figure 22.4.4B, shows an example of a prostate section hybridized with centromere probes for centromeres 7 (green) and 8 (red). Chromosomal losses have been identified when more than 50% of the nuclei exhibit a reduction of signal number, and tetraploidy has been assumed when all chromosomes investigated show signal gains up to four. For some classes of tumor, extreme polyploidy together with complex patterns of chromosomal rearrangement means that it is not realistic to select a suitable control chromosomal region in which two signals are expected. In such situations, it may be helpful to perform flow cytometric analysis of DNA content in parallel with interphase FISH analysis. Fluorescence In Situ Hybridization (FISH) Analysis of Translocation and Inversion Probes If the probes based on green and red fluorescence used for FISH are close to specific translocation breakpoints on different chromosomes, they will appear joined as a result of the translocation, generating a yellow color fusion signal. Commercial probes are now available for many of the common translocations in cancers (Table 22.4.19). One such probe from Vysis detects the Philadelphia chromosome (Ph) resulting from the 22.4.34 Supplement 23 Current Protocols in Cell Biology Table 22.4.19 Commonly Used Commercial Probes for Detection of Translocations in Sarcomas and Hematological Malignancies Neoplasm Chromosomal location Probe Scoring method CML/pediatric ALL 9q34/22q BCR/ABL Color fusion observed in metaphase and interphase Various leukemias 11q23 MLL Split signal, metaphase Various leukemias 5q31 EGFR1 Loss of signal, metaphase/interphase Various leukemias 7q31 DSS486 Loss of signal, metaphase/interphase AML M4 EO Inv(16) CBFB Split signal, metaphase Various leukemias 20q13.2 ZNF217, D20S183 Loss of signal, metaphase/interphase Various hematologic malignancies 8q24 /14q32 MYCC/IgH Color fusion observed in metaphase and interphase AML-M1 12p13/21q22; 8q22/21q22 TEL/AML1; AML1/ETO Color fusion observed in metaphase and interphase AML-M3 15q22/17q21.1 PML/RARA Color fusion observed in metaphase and interphase Ewings sarcoma t(11;22) (q24;q12) FLI1/EWS Color fusion observed in metaphase and interphase Rhabdomyosarcoma t(2;13)(q35;q14) PAX/FKR Color fusion observed in metaphase and interphase translocation between ABL on chromosome 9 and BCR on chromosome 22. Shown in Figure 22.4.4C is an example of combined G-banding and FISH analysis using the Vysis BCR/ABL probe set. Panel i of Figure 22.4.4C shows the G-banded metaphase spread to which the BCR/ABL probe was subsequently hybridized. In panel iii of Figure 22.4.4C, detection of a Ph chromosome in interphase nuclei of leukemia cells is achieved by the presence of two double-fusion (D-FISH) signals. All nuclei positive for the translocation contain one red signal (BCR gene), one green signal (ABL gene), and two intermediate fusion yellow signals because the 9;22 chromosome translocation generates two fusions, one on the 9q+ and a second on the 22q−. The following general guidelines may be helpful for performing this type of assay. 1. Green and red signals that are juxtaposed but not overlapping should be scored as ambiguous. 2. Do not score nuclei that are missing a green or red signal. This assay is looking for the presence or absence of a fusion signal, not the absence of a green or red signal. 3. Atypical signal patterns have been reported and are now considered to be clinically important (Kolomietz et al., 2001). Table 22.4.19 lists some of the commonly used FISH assays in hematological cancers as well as sarcomas. In addition to the scientific literature, readers are referred to the suppliers web sites (see Internet Resources and SUPPLIERS APPENDIX), which will provide the most up-to-date listing of currently available probes and the preferred scoring method. Inversions are related to translocations such that a break and rejoining occurs within the resident chromosome. Probes are available to detect common inversions present in AMLs Cell Biology of Chromosomes and Nuclei 22.4.35 Current Protocols in Cell Biology Supplement 23 (see Table 22.4.19). Mapping of breakpoints is common in many research cytogenetic laboratories and involves chromosome walking. To identify the locus containing the breakpoint, rough cytogenetic analysis locates the band region. Clones are then obtained spanning the putative breakpoint. These clones are then FISHed to the specimen of interest. Such is the case in Figure 22.4.4D. An inversion was identified involving the 11p15.5 region. Clones 200 kb apart, containing the IGF2 gene (green) and H19 (red) were hybridized to the patient specimen. The normal chromosome 11 shows the red and green signal hybridizing on top of each other at 11p15.5. The inverted 11 shows the clear split of signal along the abnormal chromosome 11 indicating the breakpoint lies within the 200-kb distance between 1GF2 and H19. Use of FISH Probes in Assessing Solid Tumors and Gene Amplification Gene amplification is one of the mechanisms by which cancer cells achieve over expression of some classes of oncogenes, which involves an increase in the relative number of copies of a gene per cell. This can range from one or two additional copies per cell to extreme examples where over a thousand copies per cell have been reported. Gene amplification can occur in association with the over-expression of oncogenes, thus conferring a selective growth advantage or mechanism of acquired resistance to chemotherapeutic agents leading to poor prognosis. Gene amplification is highly suited to FISH analytical approaches that have the added benefit of excellent sensitivity and the ability to address cellular heterogeneity. Neuroblastoma is characterized by the frequent occurrence of a highly amplified oncogene, MYCN. It has been known for many years that the presence of this aberration is strongly associated with poor outcome. More aggressive management is usually required when MYCN is found to be amplified. Similarly, breast cancer can be accompanied by an amplified oncogene HER2/Neu, and presence or absence of this aberration may determine which of different treatment regimens are followed. Examples of metaphase and interphase FISH assays for gene amplification are shown in Figure 22.4.4E. In this figure, a DNA probe containing the MYCN gene was hybridized to a cytogenetic specimen from a neuroblastoma patient. Panel i of Figure 22.4.4E illustrates double minute (dmns) chromosomes containing hundreds of copies of the MYCN gene as extrachromosomal bodies. This is in contrast to another patient where amplification of the MYCN gene occurs as a homogeneously staining region (HSRs) inserted in a chromosome other than the resident site of MYCN (normally at 2p24). Interphase nuclei in this image show a large patch of hybridization signal characteristic of HSRs. Some of the commonly detected aberrations observed in solid tumors which are amenable to FISH analysis are presented in Table 22.4.20. Fluorescence In Situ Hybridization (FISH) Analysis of Telomere Probes Telomeres are located at the ends of chromosomes and are characterized as (T2 AG3 ) repeat sequences and their associated proteins (Poon et al., 1999). Maintained by the ribonucleoprotein complex, telomerase, they function to protect chromosomes from endto-end fusions. In most normal tissues, telomerase is expressed at very low levels. As such, each round of DNA replication results in the gradual shortening of the telomeres. The shortening of telomeres is associated with replicative cell senescence. The up-regulation of telomerase extends the proliferative lifespan of a normal cell. In abnormal cells, expression of telomerase is associated with the maintenance of telomere length or telomere lengthening. Conventional Southern blot analysis gives average telomere length but fails to yield information on individual chromosome ends. It also underestimates the size and number of short telomeres. Although DNA probes for these sequences are available commercially, the use of commercial PNA probes for such sequences, as well as for centromeric and pancentromeric sequences, has enabled researchers to determine the overall 22.4.36 Supplement 23 Current Protocols in Cell Biology Table 22.4.20 FISH Probes Used For Assessing Solid Tumors and Gene Amplification Neoplasm Chromosomal location Probe Scoring method Neuroblastoma 2p24; 2p23-24 MYCN Interphase, metaphase; amplification Ewings sarcoma t(11;22) (q24;q12) FLI1/EWS Color fusion observed in metaphase and interphase Breast cancer 17q11.2-q12 HER2/neu Interphase amplification Glioblastoma 7p10-p21 EGF-R Interphase amplification Bladder cancer Centromere regions of Centromere 7, 13, 9 Interphase enumeration chromosomes 3, 7,17 and 9p21 region of chromosome 9 telomere lengths of individual cells (interphase nuclei) and chromosomes (metaphases) using quantitative digital imaging, as described by Poon (1999), with greater specificity and accuracy. Both fixed cytogenetic cells (Poon et al., 1999) and paraffin-embedded samples (Vukovic et al., 2003) have been used for telomere analysis. Panel i of Figure 22.4.4F illustrates subtelomeric PNA probes hybridized to a prostate cell line metaphase spread. Digital imaging and signal intensity ratios determine the relative telomere length. In some cases, loss of telomere signals can also be identified as in panel ii of Figure 22.4.4F (arrow). In this figure, a prostate cell line was hybridized with subtelomeric PNA probe as well as a PNA pan-centromeric probe. The pancentromeric probe confirmed the presence of multicentric chromosomal structures indicative of chromosomal instability. For those who wish to engage in telomere studies, access to digital imaging and analysis software capable of determining telomere length is suggested. Like all other FISH experiments, the background should be minimal. Telomere signals are relatively small and located at the ends of chromosomes. Background, such as antibody speckling can greatly affect the sensitivity of the analysis. The proper controls must also be established. Since telomere length is dependent on the number of cell divisions, age- and sex-matched controls should be included in all experiments. REAGENTS AND SOLUTIONS Use deionized or distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2A; for suppliers, see SUPPLIERS APPENDIX. Agarose gel, 2% (w/v) Dissolve 2 g molecular biology–grade agarose in 1× TBE buffer (APPENDIX 2A) by either microwaving or placing on a hot plate, and allow to cool slightly. Add 5 µl of 10 mg/ml ethidium bromide in a well vented chemical hood. Pour into casting trays (APPENDIX 3A) and allow to solidify. Store up to 1 week at 4◦ C covered with foil. AP-labeled antibody mixture Dilute anti-biotin or anti-digoxigenin conjugated to alkaline phosphatase (AP; Invitrogen) to a final concentration of 0.75 U/ml in 100 mM Tris·Cl (pH 7.5)/15 mM NaCl (APPENDIX 2A). Prepare fresh for each experiment. Cell Biology of Chromosomes and Nuclei 22.4.37 Current Protocols in Cell Biology Supplement 23 Antibodies Prepare antibodies stocks according to the manufacturer’s instructions. Tertiary antibodies are conjugated to fluorchromes. Store up to several months as frozen aliquots. Prepare working solutions of primary and secondary antibodies according to the manufacturer’s instructions using the guidelines for final antibody listed in Table 22.4.14. Diluted antibodies should be kept at 4◦ C in light-proof containers for 2 to 3 months. Minor adjustments in concentration will need to be made to account for background and signal intensity. DAPI, 100 µg/ml Dilute all of the powder from a 1-mg bottle of 4 ,6-diamidino-2-phenylindole (DAPI; Sigma) in 10 ml buffer to produce a 100 µg/ml stock solution. Store in small aliquots up to 1 year at −20◦ C. CAUTION: DAPI is a potential carcinogen and should be handed with care. DAPI in antifade Combine the following in order: 5.0 ml 1× PBS (APPENDIX 2A) 500.0 µl 100 µg/ml DAPI stock (see recipe; 1 µg/ml final) 0.5 g p-phenylenediaminie (Sigma; 10 mg/ml final; dissolve well) 45.0 ml glycerol (90% v/v final) Transfer to a 50.0-ml conical tube, wrap with aluminum foil (product is light sensitive), and place on a rotator 30 min to ensure proper mixing. Store in 1-ml aliquots up to 1 year at −20◦ C. The resulting solution is very viscous This reagent is available commercially as Vectashield (Vector Laboratories). DNase I, 3 mg/ml 3.0 mg DNase I powder 500.0 µl glycerol (50% v/v final) 50.0 µl 1 M Tris·Cl, pH 7.5 (50 mM final; APPENDIX 2A) 5.0 µl 1 M MgCl2 (5 mM final) 1.0 µl 1 M 2-mercaptoethanol (1 mM final) 1.0 µl 10 mg/ml BSA (10 µg/ml final) Adjust volume to 1.0 ml with H2 O Store in 50-µl aliquots up to 1 year at −20◦ C. Fluorescence In Situ Hybridization (FISH) DNase I dilution buffer 250.0 µl 1 M Tris·Cl, pH 7.0 (50 mM final; APPENDIX 2A) 25.0 µl 1 M MgCl2 (5 mM final) 5.0 µl 1 M 2-mercaptoethanol (1 mM final) 20.0 µl 10 mg/ml BSA (4 µg/ml final) 4.7 ml H2 O Store in 1-ml aliquots up to 6 months at −20◦ C or In 1 ml aliquots up to 1 month at 4◦ C. 22.4.38 Supplement 23 Current Protocols in Cell Biology dNTP mixture, 1 and 2 mM For a 1 mM mixture: 1.0 µl 100 mM dATP 1.0 µl 100 mM dCTP 1.0 µl 100 mM dGTP 97.0 µl H2 O, sterile Store in 100-µl aliquots up to several months at −20◦ C For a 2 mM mixture: Double the volume of each 100 mM dNTP added (i.e., use 2 µl of 100 mM dNTP) and reduce the amount of water to 94 µl. Store up to several months. The total volume of either solution is 100 µl. Fluorochrome/dTTP mixture The amount of fluor-dUTP will vary. Thus, the recipes below are guidelines. Refer to the manufacturer’s suggested final concentrations. All labeled nucleotides should be stored in 20 to 30-µl aliquots up to 6 months at −20◦ C in light-proof containers. FITC-dUTP/dTTP: 3.5 µl 1 mM dTTP (0.6 mM final) 1.75µl 1 mM fluorescein (FITC)-dUTP (0.3 mM final; Roche) The total volume, 5.25 µl, is appropriate for one labeling reaction and can be scaled up as needed. Rhodamine-dUPT/dTTP mixture: 3.5 µl 1 mM dTTP (0.6 mM final) 1.75 µl 1 mM rhodamine-dUTP (0.3 mM final; Roche) The total volume, 5.25 µl, is appropriate for one labeling reaction and can be scaled up as needed. Texas Red-dUPT/dTTP mixture: 3.5 µl 1 mM dTTP (0.7 mM final) 1.0 µl 1 mM Texas Red-dUTP (0.2 mM final; Roche) The total volume, 4.5 µl, is appropriate for one labeling reaction and can be scaled up as needed. Cy3-dUPT/dTTP mixture: 3.5 µl 1 mM dTTP (0.7 mM final) 1.25 µl 1 mM Cy3-dUTP (0.2 mM final; Amersham Biosciences) The total volume, 4.75 µl, is appropriate for one labeling reaction and can be scaled up as needed. Cy5-dUPT/dTTP mixture: 3.5 µl 1 mM dTTP (0.7 mM final) 1.25 µl 1 mM Cy5-dUTP (0.2 mM final; Amersham Biosciences) The total volume, 4.75 µl, is appropriate for one labeling reaction and can be scaled up as needed. Cell Biology of Chromosomes and Nuclei 22.4.39 Current Protocols in Cell Biology Supplement 23 Formamide, 70%/Tris·Cl (pH 7.0 to 7.5), 10 mM/BSA, 0.1% (w/v) Dissolve 100 mg BSA in 29 ml water. To this solution, add 70 ml formamide (Invitrogen) and 1 ml of 1 M Tris·Cl, pH 7.0 to 7.5 (APPENDIX 2A). Store up to several weeks. CAUTION: Formamide is a carcinogen and should be handled with care. Discard according to institution’s biohazard rules. Total volume is 100.0 ml; however, the solution can be made in larger quantities and stored up to several weeks at 4◦ C. Hapten/dTTP mixture The amount of hapten-dUPT will vary. Thus, the recipes below are guidelines. Refer to the manufacturer’s suggested final concentrations. Store up to several months or a year at −20◦ C. Biotin-dTTP/dUTP mixture: 3.5 µl 1 mM dTTP (0.6 mM final) 1.75 µl 1 mM biotin-16dUTP (0.3 mM final; Invitrogen) The total volume, 25 µl, is appropriate for one labeling reaction and can be scaled up as needed. Digoxigenin-dTTP/dUTP mixture: 3.5 µl 1 mM dTTP (0.6 mM final) 1.75µl 1 mM dig-11dUTP (0.3 mM final; Roche) The total volume, 5.25 µl, is appropriate for one labeling reaction and can be scaled up as needed. Hybridization buffer 500.0 µl high grade formamide (50% v/v final; Invitrogen) 100.0 µl 20× SSC (2× final; UNIT 18.6) 100.0 µl dextran sulfate (10% final) 300.0 µl H2 O Store in 100-µl aliquots up to several months at 4◦ C Total volume is 1.0 ml. Hybridization buffer can also be purchased from DAKO. Loading dye, 5× 0.125 g bromphenol blue (0.25% final) 15.0 ml glycerol (30% final) 35.0 ml H2 O Store in 1.0-ml aliquots up to several months at 4◦ C Total volume is 50.0 ml. Fluorescence In Situ Hybridization (FISH) NBT/BCIP 22.5 µl 75 mg/ml NBT (Invitrogen) 17.5µl 50 mg/ml BCIP (Invitrogen) 4.96 ml 100 mM Tris·Cl (pH 9.5; APPENDIX 2A)/100 mM NaCl/50 mM MgCl2 Prepare fresh for each experiment. Volumes can be scaled up or down as required. 22.4.40 Supplement 23 Current Protocols in Cell Biology Nick translation buffer, 10× 1.0 µl 10 mg/ml BSA (0.1 µg/µl final) 10.0 µl 1 M 2-mercaptoethanol (0.1 M final) 50.0 µl 1 M Tris·Cl (0.5 M final) 5.0 µl 1 M MgCl2 (50 mM final) 34.0 µl H2 O Store in 100-µl aliquots up to several months at 4◦ C Total volume is 100.0 µl. PBS, 1×/MgCl2 , 0.05 M 950.0 ml 1× PBS 50.0 ml 1 M MgCl2 Store at room temperature until ready for use. The total volume is 1 liter. Pepsin, 0.5% (w/v) in NaCl, 0.85% Dissolve 500 mg pepsin in 100.0 ml of 0.85% (w/v) sodium chloride. Adjust pH to 1.5 with 12 N HCl. Prepare fresh for each experiment. PI, 100 µg/ml Dissolve the powder from an entire 10-mg bottle of propidium iodide (PI; Sigma) in 10 ml water to a final concentration of 100 µg/ml. Store in small aliquots indefinitely at −20◦ C. PI in antifade Combine in the following order: 5.0 ml 1(PBS; APPENDIX 2A) 150.0 µl 100 µg/ml PI (0.3 µg/ml final; see recipe) 0.5 g p-phenylenediaminie (10 mg/ml final; Sigma; dissolve well) 45.0 ml glycerol (90% final) Transfer to a 50.0-ml conical tube, wrap in aluminum foil (the product is light sensitive), and place on a rotator 30 min to ensure proper mixing. Store in 1-ml aliquots up to several months at −20◦ C. The resulting solution (50 ml total) is very viscous. This solution is available commercially available from Vectashield (Vector Laboratories). RNase I, 100 µg/ml Prepare a final 100 µg/ml solution of RNase I in 2× SSC (UNIT 18.6) fresh for each experiment using any molecular-grade RNase enzyme. Sonicated salmon sperm DNA standards Prepare 50.0 ng/µl salmon sperm DNA standard by combining 5.0 µl of a 10 µg/µl stock (Invitrogen), 300 µl of 5× loading dye (e.g., UNIT 18.6), and 695 µl water. Prepare a 25.0 ng/µl standard by combining 500 µl of the 50 ng/µl standard, 100 µl of 5× loading dye, and 400 µl water. Prepare a 12 ng/µl standard by combining 500 µl of the 25.0 ng/µl standard, 100 µl of 5× loading dye, and 400 µl water. Store all standards up to several months at 4◦ C or indefinitely at −20◦ C. The average size of the commercial salmon sperm is ∼500 to 2.0 kb Cell Biology of Chromosomes and Nuclei 22.4.41 Current Protocols in Cell Biology Supplement 23 Triethanolamine (pH 8.0), 0.1 M 660.0 µl triethanolamine (final 0.1 M; Sigma) 50.0 ml H2 O Prepare fresh for each experiment. Total volume is 50.0 ml. COMMENTARY Background Information Fluorescence In Situ Hybridization (FISH) Applications of FISH As early as 1988 (Lichter et al., 1988), FISH was used to visualize labeled DNA probes hybridized to chromosome and interphase nuclei preparations. The improvement of cloned DNA sources, antibodies, fluorochromes, microscopy and imaging equipment, and software has permitted a variety of scientific investigations. FISH analysis is routinely used in clinical cytogenetic laboratories for detection of syndromes in prenatal assessments, including Prader-Willi/Angelman Syndrome, DiGeorge Syndrome, and Cri-du-chat. Cancer cytogenetics laboratories use FISH to confirm and monitor hematological malignancies such as CML and AML (Table 22.4.19 and Table 22.4.20). Gene amplification studies for neuroblastoma (MYCN amplification), and breast cancer (HER2/NEU amplification) are also routinely carried out. Mapping of newly identified genes have relied heavily on FISH (Squire et al., 1993; Lichter et al., 1988) both in humans and other species (Boyle et al., 1990, Giguere et al., 1995). Metaphase spreads may not always be easy to obtain, thus interphase cells offer a means of obtaining information, albeit via an indirect method. This has been useful for gauging chromosome instability (Speicher et al., 1995; Ghadimi et al., 1999; Al-Romaih et al., 2003; Vukovic et al., 2003) and determining normal versus abnormal content, as well as gene amplification or deletion. FISH applied to paraffin-embedded sections appeared in the early 1990s (Thompson et al., 1994) and was applied to both thick (>5 µm) and thin (5 µm) paraffin sections. Application of FISH to paraffin-embedded sections provides a tremendous opportunity to correlate the histopathological classification to the genomic changes detected. Furthermore, it enables the investigator to study concepts of cellular heterogeneity, tumor focality, and metastasis (Squire et al., 1996). Paraffin FISH analysis requires patient and careful technical expertise. Proper controls must be used when interpreting final results. Depending on the type of information sought, paraffin FISH is an acceptable form of data collection when fresh tissues for cytogenetic suspensions are not available; however, paraffin FISH will not provide chromosome-based information. Preparation of probes The development of reliable cloning strategies in the 1980s facilitated the genomic analysis and sequencing of specific DNA fragments. Mapping of these genes to their chromosomal locations was previously laborious and infrequently reliable. The emergence of FISH in the early 1990s paved the way for an effective and direct means of mapping specific DNA fragments to their chromosomal locations. Types of probes. The creation of comprehensive genomic libraries as a result of the human genome project provides renewable resources for FISH probes. At present, bacterial artificial chromosomes (BACs) are the most popular cloned forms of genomic DNA used for FISH probes. Although yields are generally considered low in comparison to plasmids and cosmids (predecessors of BACs), BAC inserts are larger (200 kb) and can produce a stronger FISH signal compared to cosmids and plasmids, which are considerably smaller in insert size (i.e., ranging from 2 to 30 kb). P1 artificial chromosomes (PACs) and yeast artificial chromosomes (YACs) have also been used in the past and are still used in many research laboratories. Other probes, such as RNA and oligonucleotide probes are options and are reviewed in Speel (1999), and Schwarzacher and Heslop-Harrison (2000). More recently, peptide nucleic acid (PNA) probes (Lansdorp, 1996), have been used for FISH experiments with great success (Martens et al., 1998; Vukovic et al., 2003.). Using the same nucleotide bases as DNA probes, PNA probes do not have a phosphate or deoxyribose sugar backbone. Specific fluorochromes, haptens, or enzymes can be chemically attached to the bases and used in the same fashion as DNA probes. PNA probes are sequence specific. Thus, sequence information must accompany a PNA custom probe 22.4.42 Supplement 23 Current Protocols in Cell Biology order to companies such as Applied Biosystems (http://www.appliedbiosystems.com). There are several advantages of PNA probes. For instance, PNA has a neutral backbone, which provides stronger binding and greater specificity of interaction. Also, the uncharged PNA structure creates stronger binding independent of salt concentration, allowing more robust hybridization applications. Finally, the fact that PNA oligomers have resistance to nucleases and proteases, permitting more robust in situ experiments, is also an advantage. The drawback of PNA probes are in the cost of their fabrication. The typical molecular (cytogenetic) laboratory cannot generate these probes in-house. The cost-effectiveness of PNA probes, therefore, can only be appreciated if the laboratory is carrying out a particular hybridization frequently, as in the case of a clinical cytogenetic laboratory. Methods for preparing probes. By far the most commonly used strategy for labeling DNA probes is by nick translation. Nick translation involves the simultaneous actions of two enzymes: DNase I and DNA polymerase I. DNase I randomly nicks the DNA fragment in each strand of the double-stranded DNA molecule. DNA polymerase I (derived from E. coli), with its three activities—i.e., exonuclease function (removes bases in the 5 to 3 direction), polymerase function (adds nucleotides from the 3 nick site), and the 3 to 5 proof reading function—incorporates the label, whether indirectly using a hapten (i.e., biotin or digoxigenin) or directly (i.e., fluorescein or rhodamine). Nick translation can be applied to all cloned DNA sources without the need to remove vector sequences. Polymerase chain reaction (PCR) is another commonly used method of labeling DNA probes from cloned sources, as well as from microdissected genomic DNA or flow-sorted chromosomes. PCR employs multiple rounds of template denaturation, primer annealing, and template replication, facilitated by Taq DNA polymerase. Primers are variable and may include sequence-specific primers to amplify those targeted fragments, vector sequence primers to amplify and label cloned DNA, and/or universal primers. Compared to nick translation, which requires micrograms of starting DNA, PCR requires only nanograms of template. Furthermore, nick translation is most efficient when the starting DNA is of high molecular weight with little or no degraded material. PCR labeling is a little more forgiving of DNA quality; thus, suboptimal DNA may still be used when not acceptable for nick translation. De- pending on the template, several rounds of amplification may be required before labeling, as in the case of in-house production of chromosome painting probes. Other labeling strategies, including random priming and endlabeling, are also options and are described in Schwarzacher and Heslop-Harrison (2000). For both nick translation and PCR labeling, commercial kits and enzyme mixes are readily available and optimized. Usually, commercial kits combine DNase I and DNA polymerase I as an enzyme mix. The drawback of combining them is the fact that one enzyme (usually the DNA polymerase I) will lose its enzymatic activity before the other. Commercial kits can also be costly. Making an in-house kit allows the investigator to alter enzyme concentrations as necessary to better optimize the reaction. This alleviates the combined-enzyme problem with commercial kits. In-house kits are also expensive and they do require rigorous internal quality control and troubleshooting from time to time. See Critical Parameters for discussion of slide preparation, hybridization, posthybridization washes and detection, and interpretation of FISH findings. Critical Parameters Preparation of probes DNA quality. Ensure that the starting DNA to be labeled has a high molecular weight with no degraded DNA fragments or protein. Contaminating RNA will contribute slightly to the spectrophotometric quantification of the DNA and cause a miscalculation of the actual starting amount of DNA. DNA should be assessed using an accurate spectrophotometer and by gel electrophoresis. Compare the concentration of DNA as determined using a spectrophotometer and gel with a DNA of known concentration. The DNA used as a standard for concentration can be another undigested clone or genomic DNA. The starting DNA should be dissolved in water rather than Tris/EDTA (TE) buffer. TE is normally used to inhibit any DNases that may be present in the water. By that reasoning, TE buffer has been known to inhibit the enzymatic activity of the DNase during the nick translation reaction. Furthermore, it may inhibit the polymerase activity for nick translation and PCR labeling. Reagents. Ensure that reagents are used prior to their expiration dates. The most critical reagents in the labeling reactions are: Cell Biology of Chromosomes and Nuclei 22.4.43 Current Protocols in Cell Biology Supplement 23 1. DNA polymerase I/Taq DNA polymerase. Failure of polymerase activity in the reaction will lead to inefficient incorporation of the labeled nucleotide into the DNA, resulting in a very weak FISH signal. 2. Conjugated nucleotides. Nucleotides conjugated to a fluorochrome are light sensitive. Repeated and prolonged exposure to direct light will quench the fluorescent signal. Store these reagents in small aliquots at −20◦ C in light-proof containers. Restrict exposure to light during the labeling procedure and store labeled DNA at −20◦ C in lightproof containers. Unlike nonfluorescently labeled probes, these directly-labeled DNAs will have a shorter shelf life. The concentration of nucleotides can vary from as much as 1 mM to as low as 0.2 mM in different nick translation protocols. The labeled nucleotide should always be in at least a 2- fold excess of the corresponding unlabeled nucleotide. Furthermore, the labeled nucleotide can be interchangeable between hapten-dUTP or hapten-dATP. If the investigator chooses to use Biotin 14-dATP rather than the dUPT form, the nucleotide mixture should be adjusted accordingly. 3. 2-Mercaptoethanol. Buffers containing 2-mercaptoethanol should be replaced every 3 to 4 months, particularly if not stored at −20◦ C. 4. DNase I. Although the activity of DNase I is usually potent enough that it may be used for up to a year when stored at −20◦ C, the specific activity may decrease over time. This is evident when the fragment sizes are larger than expected when the standard DNase I concentration and incubation times are used. Simply increase the amount of DNase I used in the reaction and keep the same period of incubation, or keep the same amount of DNase I used in the reaction and increase the time of incubation. 5. PCR factors. Treat PCR labeling like any other PCR experiment and consider the following factors: amount of template, pH, magnesium ion concentration, and primer sequence. Equipment. Small volumes are used for both labeling strategies. Pipets should be calibrated and serviced often. Use an accurate spectrophotometer and water bath that can effectively maintain a constant temperature, if a thermocycler is not available for nick translation. Fluorescence In Situ Hybridization (FISH) Preparation of slides FISH involves the hybridization of the labeled and denatured DNA probe to its denatured DNA target located on the slide. Each component requires care to obtain the most effective results. Pretreatment of the slide with a protease before denaturation permits the removal of any cellular and cytoplasmic debris. In the case of paraffin-embedded sections, more aggressive treatment is required to remove residual wax as well as to allow sufficient protease treatment to permit access of the probe to the intended target DNA. Protease treatment. Treatment with pepsin has been featured in this protocol because its action is relatively mild as compared to other proteases such as proteinase K; however, many laboratories use proteinase K alone or in combination with pepsin, particularly for the digestion of paraffin-embedded sections, to obtain the proper digestion conditions for FISH (see Table 22.4.9). The incubation times of any protease treatment will vary according to the type of tissue (e.g., paraffin-embedded), quality of the slide (i.e., quantity of cytoplasmic debris/cellular debris), as well as the lot or batch of pepsin used. It is suggested that incubation times be increased rather than the concentration of pepsin used in the digest. The temperature at which the incubations occur is also critical. Higher temperatures facilitate faster digestions as opposed to those at room temperature or below. While variations in the pretreatment protocol can be made, the starting quality of the fixed tissue is the most critical factor. Ideally, the tissue should be fixed shortly after removal from the host. Paraffin-embedded samples. In the case of paraffin-embedded tissues, formalin is the fixative of choice for routine use in most clinical laboratories; however, superior results for most research needs may be obtained if tissue sections are fixed using ethanol or paraformaldehyde prior to embedding. Buffered formalin solutions have also been implemented to provide better recovery of DNA, RNA, and protein for future studies. The time spent in fixation is equally as important as the type of fixative. The longer the time spent in fixative, the more difficult it may be to denature the DNA to a singlestranded state. This is due to the cross-linking action of the fixative to proteins that enables the cells to maintain their morphology. Sections for FISH should be cut fresh from the original block, preferably a few sections from the exposed face. This will help to ensure that the DNA has not been exposed to DNases, or oxidative damage from the handling of the block or from the local environment. Several sections should be taken so that different pretreatment conditions can be tested. It is suggested that the investigator take the time to contact the 22.4.44 Supplement 23 Current Protocols in Cell Biology histology laboratory preparing the specimens to document the fixation procedure for troubleshooting later. Denaturation. Denaturation is also a critical parameter for successful FISH. Too much denaturation of the target DNA causes excessive DNA damage, yielding poor banding and/or hybridization. Too little denaturation results in ineffective hybridization due to limited access to the target DNA. The extent of denaturation will depend on several factors. 1. Quality of the slide. Namely, the presence of cytoplasmic and/or cellular debris. 2. Age of the slide. Whether using a fresh slide, a fresh slide with artificial aging, a slide that is 1 week or 1 month old, and so forth. 3. Previous assays. Namely if banding has previously been performed on the slide. 4. Tissue type. Whether fibrous, bony, or muscle tissue, or the like, is being used. 5. Age and method of fixation. See above. 6. Section thickness. Normally 5-µm-thick sections are used. Hybridization Optimal hybridization is influenced by many factors that will affect the target DNA and the probe itself. These factors are sources for inefficiencies and should be carefully monitored and documented such that effective troubleshooting can be carried out. Stringency. The hybridization of a DNA probe to its DNA target is dependent on the extent of sequence homology, the complexity of the DNA sequence, and the chemical factors that influence the success of maximal binding efficiency. Stringency refers to the percentage of matches and mismatches between the probe and target such that the hybridized double helix remains stable. FISH experiments are typically run at 70% to 90% stringency, with 70% representing a lower relative stringency than 90%. This is critical when determining the feasibility of carrying out cross-species FISH experiments (i.e., human probes on mouse targets or vice versa), where sequence homology may be considerably lower in parts of the probe. The reagents present in hybridization buffers, as well as the temperature significantly influence stringency. Temperature. DNA strand melting and annealing are influenced by temperature. Since reagents such as formamide are present in the hybridization buffer, high temperatures (i.e., 80◦ to 90◦ C) are not required for DNA denaturation. This is the reason denaturation of the DNA can be accomplished at 72◦ C in the presence of formamide. Increasing temperatures also increases stringency, such that a 1◦ C alteration increases the stringency by 1%. Thus, increasing the hybridization temperature from 37◦ to 42◦ C will increase the stringency of the hybridization step, requiring the probe to possess higher sequence homology with its target. This may be particularly useful if there is a chance of crosshybridization, which is typically the case with locus-specific probes of genes that belong to a class of genes/sequences that are very similar. Pseudogenes may also cross-hybridize. Lowering the hybridization temperature under 37◦ C will result in lower stringency conditions permitting the hybridization of the probe to targets with greater mismatch. This also has advantages and disadvantages. At low stringency, mismatched spurious hybridization events will be more common and contribute to more experimental noise at the detection step. Probe size. The rate of hybridization is also influenced by probe size. Generally, longer fragments (>500 bp) will require a longer time to hybridize than shorter ones. Once again, the hybridization is affected by the extent of sequence homology. PNA probes. In the case of PNA probes, the specificity of the probe, as a required function of their fabrication through oligosynthesis, eliminates some of the complexities experienced by DNA probes. Since the PNA probe has an uncharged backbone, there is no charge repulsion when the probe hybridizes to the DNA, thus creating a stronger bond. Furthermore, PNA/DNA bonds will melt 15◦ C above DNA/DNA bonds, almost independently of salt concentration. The specificity of the PNA/DNA bond lies in the increased destabilization of the PNA/DNA bond when a mismatch occurs. Post-hybridization washes and detection Stringency. The factors that affect hybridization also influence the conditions involved in post-hybridization washes and antibody incubations. Incubation at 37◦ C under a range of 50% to 60% formamide in 2× SSC sets a stringency of ∼70% to 80%. This permits hybridization of sequences with a 20% to 30% mismatch and maintains duplex stabilization. Increasing hybridization temperatures raises the stringency, requiring greater sequence homology for effective hybridization. Conversely, decreasing temperatures permits hybridization with greater mismatch. Decreasing salt ion strength (i.e., SSC) also increases Cell Biology of Chromosomes and Nuclei 22.4.45 Current Protocols in Cell Biology Supplement 23 Table 22.4.21 Stringency of DNA/DNA Hybridization and Post-Hybridization Washesa Stringency as a function of % formamide SSC concentration Temperatureb 60 55 50 45 40 35 30 5× 37◦ C 76 73 70 67 64 61 58 2× ◦ 37 C 83 80 77 74 71 68 65 1× ◦ 37 C 88 85 82 79 76 73 70 0.75× 37◦ C 90 87 84 81 78 75 72 0.5× 37◦ C 93 90 87 84 81 78 75 0.2× 37◦ C 100 96 93 90 87 84 81 0.1× 37◦ C 105 101 98 95 92 89 86 5× 42◦ C 81 78 75 72 69 66 63 2× 42◦ C 88 85 82 79 76 73 70 1× ◦ 42 C 93 90 87 84 81 78 75 ◦ 42 C 95 92 89 86 83 80 77 0.5× ◦ 42 C 98 95 92 89 86 83 80 0.2× ◦ 42 C 105 101 98 95 92 89 86 0.1× ◦ 110 106 103 100 97 94 91 0.75× 42 C a This Table describes the changes in stringency for probes with 43% GC content at 300 bp (modified from Schwarzacher and Heslop-Harrison, 2000). Increasing the formamide content or temperature, or decreasing the SSC strength will increase stringency. A combination of these three factors can be used to modulate the stringency. b Hybridization of probe to DNA typically occurs under 50% formamide in 2× SSC at 37◦ C, giving a 77% stringency. Washing of unbound probe following hybridization typically occurs under 50% formamide in 2× SSC at 42◦ C, giving an 82% stringency. Fluorescence In Situ Hybridization (FISH) stringency. The combination of temperature and salt concentration can affect hybridization kinetics considerably. The same holds true for removal of unbound probe. Typically, post-hybridization washes are carried out at 42◦ to 45◦ C. This temperature, coupled with 50% formamide and 2× SSC, permits 90% stringency for probes with a 43% GC content, at a probe size of 300 bp (Schwarzacher and Heslop-Harrison, 2000). Table 22.4.21 describes the stringency conditions based on these parameters. Detergents. Some suppliers of commercial probes may suggest higher temperature washes of 72◦ C in a low ion (high stringency) SSC wash containing a detergent. This too is based on high sequence homology and the relative GC sequence content of the probe. This rapid wash method eliminates the need for formamide. Caution should be exercised when using high temperatures, since there is the risk of partial denaturation of the hybridized probe and target. This will result in loss of signal. A rapid wash should only be used with probes that possess high sequence homology and high labeling efficiency. Commercial probes tend to fit these criteria. The subsequent SSC washes and washes with detergents serve to remove any remaining unbound probe and antibodies. As before, decreasing the SSC content will create a more stringent wash. The choice of detergent is subjective, largely based on the preference and experience of the investigator. The most commonly used detergents include Tween 20, NP-40, IPEGAL, SDS, and Triton X-100 (reviewed in Speel, 1999). In general, the actions of these detergents are comparable when used at the standard 0.1% in 4× SSC; however, SDS, Triton X-100, and NP-40 are generally harsher than Tween 20. If high background persists, the number of washes with the detergent can be increased or the percentage of detergent and SSC strength can be altered. Antibodies. The choice of antibody systems is also fairly standard. Many companies provide antibodies raised in different animals and conjugated to different fluorochromes so that a variety of probes may be used and detected at one time. Some antibodies may require a higher concentration; thus, it is important that the signal strength be monitored carefully for background and the antibody concentration adjusted as required. As discussed 22.4.46 Supplement 23 Current Protocols in Cell Biology above, caution should be exercised when using various antibody combinations for detecting more than one probe: ensure that there is no cross-reactivity between antibodies raised in different species. Counterstains. Finally the use of a counterstain helps to visualize the cells or spreads. DAPI stains DNA blue upon fluorescence and is most commonly used. DAPI, however, may obscure a green signal if using a dual filter (i.e., FITC/DAPI), so a red counterstain, such as propidium iodide (PI), may be more appropriate. If the color of probe detected is red, do not use PI as a counterstain. The signals are protected from rapid quenching through the reagents used in the mounting medium. These counterstains in mounting (antifade) medium can be purchased commercially. If the counterstain is prepared in the laboratory, be sure to use high quality glycerol, since some lower quality glycerols can cause autofluorescence. The concentration of counterstain may require adjustment. Higher counterstain concentrations may obscure FISH signals. If this is the case, dilute the counterstain in antifade by adding more glycerol and PBS in amounts that will be consistent with the final concentrations. Storage. All slides should be stored at −20◦ C for long-term storage (i.e., up to 1 year) and at 4◦ C for short-term storage (i.e., up to 6 months). Troubleshooting Preparation of probes See Table 22.4.22 for a troubleshooting guide to labeling. Preparation of slides Cytogenetic specimens. The influence of pretreatment and denaturation cannot be readily assessed until the entire FISH protocol has been carried out and the slides are visualized by fluorescence microscopy (Henegariu et al., 2001). The slides, however, may be visualized by phase-contrast microscopy for cytogenetic preparations after the dehydration step following pepsin treatment to determine the extent of digestion. The slides may be returned for digestion if the cytoplasmic debris does not appear to have been removed. Similarly, for treatment of paraffin sections, following visualization for autofluorescence, the slides may be returned for digestion and reassessed. If there are no cells remaining on the slide following pepsin treatment, then the treatment was too harsh. If many cells have lifted off, the treatment was too harsh. Adjust the time or pepsin concentration as required. Paraffin sections. The diameter of nuclei in various tissue types will vary between 4 and 8 µm. The estimated diameter for the tissue of interest should be ascertained from the literature and sections of the same thickness or 2 to 4 µm greater should be cut, so that it is likely that the entire nucleas is included in the section. Pepsin digestion works for most tissue types. Occasionally, the tissue will exhibit persistent green autofluorescence of nuclei and no signal (see Table 22.4.19). Variation in histology fixation procedures and quality control performed by the hospital pathology laboratory necessitates careful assessment of treatment times and/or concentration of pretreatment (also see Table 22.4.9). Certain tissue sections are more resistant to effects of protein digestion than others. This may be due to the tissue type or amount of protein crosslinking elicited by excessive formalin fixation. If surplus fresh tissue can be obtained, then ethanol or paraformaldehyde fixation will cause less cross-linking and provide better results for most research applications. It should also be noted that normal tissues are often more refractory to protein digestion than tumor material derived from the same tissue type. If there are a limited number of slides remaining from the patient sample of interest it is sometimes helpful to use a superfluous slide from another sample of the same age and cell type to derive the optimal digestion parameters with respect to aging and storage. More comprehensive guidelines for improved paraffin results are summarized by Hyytinen et al. (1994). Anticipated Results Preparation of probes Successful labeling for FISH will yield fragment sizes between 200 and 500 bp. Fragments up to 1 kb are acceptable, but larger fragments will contribute to background. Investigators should expect that the actual amount of DNA aliquoted onto the gel for visualization should be fairly close to the expected amount loaded onto the gel. The labeling reaction is fairly tolerant to variations in concentration from half the actual amount to two-times greater than the actual amount. Once the labeling conditions have been established and tested in control FISH experiments, the investigator can be confident that subsequent labeling experiments will proceed without incident. Minor adjustments must Cell Biology of Chromosomes and Nuclei 22.4.47 Current Protocols in Cell Biology Supplement 23 Table 22.4.22 Troubleshooting Guide to FISH Probe Labelinga Problem Cause Solution DNA failed to digest DNase I not present Ensure that DNase I was added to the labeling reaction. Loss of DNase I activity If DNase I was added, check the expiration date or change to a fresh aliquot. Fragments too small TE buffer used to dissolve DNA (EDTA inactivates DNase I) If TE buffer was used to resuspend the DNA, reprecipitate the DNA, resuspend in water, quantify by spectrophotometry, and relabel. Too much DNase I added Pipetting errors are the leading cause of many experimental problems. Use accurate pipettors. DNA starting Refer back to spectrophotometer readings concentration inaccurate and gel images of undigested DNA to determine whether there were any discrepancies. Contaminating RNA and proteins may contribute to an inaccurate reading, such that the actual amount of starting DNA was less than expected, thus changing the labeling (digestion) kinetics for a specified amount of DNA over a specific time at a specific DNase I concentration. Relabel and increase the amount of starting DNA as required Fragments too long Fluorescence In Situ Hybridization (FISH) Starting DNA was degraded If the starting DNA already contains degraded fragments, they will simply continue to degrade into relatively smaller fragments. Refer back to the gel of the undigested DNA and determine the percentage of degraded fragments present. Compensate for the fragments that will be digested away and increase the amount of DNA required to relabel. Consider PCR labeling strategies or obtain a better DNA sample. Labeling time too long Be sure that the correct incubation time was used. Unstable water bath temperature The labeling reaction has been optimized for a specific temperature. Increasing the temperature will facilitate faster nicking, while decreasing the temperature will slow it. Insufficient DNase I Use accurate pipettors (see above). DNA starting As described above, check the concentration inaccurate spectrophotometer reading and gel image. If too much DNA was added to the labeling reaction, the same amount of DNase I used to nick a given amount of DNA in a specific time will increase when there is more DNA present. The labeling of this DNA can be salvaged by continuing to incubate for 20 to 30 min (or as required). Be sure to spike the reaction with fresh enzymes. continued 22.4.48 Supplement 23 Current Protocols in Cell Biology Table 22.4.22 Troubleshooting Guide to FISH Probe Labelinga , continued Problem Cause Solution Starting DNA was dissolved in TE buffer Check whether the starting DNA was dissolved in TE as this will inhibit the nicking of DNA. The labeling of this DNA can be salvaged by continuing to incubate as required. Be sure to spike the reaction with fresh enzyme. Decreasing activity of DNase I Over time, the specific activity of DNase I will decrease. If this is the case, increase the amount of DNase I used in the reaction, while keeping the same incubation period. Otherwise, keep the same DNase I concentration and increase the labeling time. No fragments present Absence of DNA added Make sure that DNA was added to the to reaction reaction Insufficient DNA added As discussed above, check to see that the concentration of DNA is accurate and pipettors are accurate. Starting DNA degraded If a significant portion of the starting DNA was degraded to begin with, as shown by gel electrophoresis, this will cause an increase in fragmentation speed, with the smaller fragments running off the gel. A new DNA specimen should be used, or the investigator should consider PCR methods. Too much DNase I added Expected amount of DNA starting DNA too low/too high concentration inaccurate (but fragmented to the right size) PCR failed Check that the proper DNase I concentration and/or volume was used in the labeling reaction. Assess using a concentration standard. If the actual amount of DNA is less than expected, as determined by gel electrophoresis, then the starting amount of DNA was insufficient or caused by some of the factors described above. If the investigator chooses to relabel, increase the amount of starting DNA as required. If the actual amount of DNA is greater than expected, again as determined by gel electrophoresis, then the starting amount of DNA was incorrect. The investigator may choose to proceed and use this probe for FISH, but the overall incorporation of label into the DNA will be lower since the labeling reaction contains sufficient dNTPs for a specified DNA quantity. If the investigator chooses to relabel, decrease the amount of starting DNA material as required. Amount of template, Refer to APPENDIX 3F and APPENDIX 2A. pH, or MgCl2 concentration incorrect, or quality of template poor a The guide assumes that the labeling conditions have generally been optimized but occasionally anomalies occur. Cell Biology of Chromosomes and Nuclei 22.4.49 Current Protocols in Cell Biology Supplement 23 be made when using new reagents. Once a probe has been FISHed, the signal should be clearly visible with little or no background or cross-hybridization. The investigator should be aware that other factors, such as hybridization efficiency, slide quality, antibody quality, stringency of post-hybridization washes can also affect the signal strength. Preparation of slides Optimal pretreatment of cytogenetic or paraffin-embedded slides should produce slides that are relatively free of cytoplasm/cellular debris, free of paraffin, and have adequately denatured DNA for FISH. The samples should not be autofluorescent. Hybridization Due to the multistepped nature of FISH, it is difficult to determine the success of hybridization until the entire assay is completed. Successful hybridization will depend on adequate denaturation of the target DNA on the slide, as well as adequate denaturation of the probe. Proper suppression of repetitive sequence elements with Cot-1 DNA can increase the specificity of hybridization. The specificity of hybridization is also regulated by stringency factors described above (see Background information; also see Critical Parameters). Successful hybridization will allow the identification of chromosomal location of the probe sequence. Post-hybridization washes and detection The protocols described provide a basic framework for adjusting post-hybridization washes and detections. Under ideal conditions, the stringency of the post-hybridization washes is sufficient to adequately remove any unbound/nonhybridized probe from the target DNA. The specificity of the probe and the suppression of repetitive elements during preannealing will also decrease the number of crosshybridization signals. The concentration of detergent used in the washes is also optimized to remove any unbound antibodies, although minor modifications, as required, should be made when using a different brand, lot, or type of antibody. The result, upon visualization with a microscope, is a clean preparation showing minimal background and an easily visualized signal(s) on the DNA target. Time Considerations Fluorescence In Situ Hybridization (FISH) Preparation of probes The most time consuming portion of labeling for FISH is in the optimization of the la- beling procedure if using an in-house protocol. As discussed previously, commercial kits for labeling are available and give relatively consistent results. Preparation of slides The preparatory work for preparing cytogenetic slides requires minimal time and the protocol is fairly robust. Optimization for paraffinembedded sections requires more careful time and trial-and-error. The preparation of cytogenic slides for FISH can be accomplished in ∼1 hr. Once optimized conditions have been obtained for paraffin-embedded tissues, it too can be accomplished in 1 to 2 hr. However, initial optimization may require several experimental attempts. Hybridization The hybridization step of FISH typically occurs overnight (i.e.,. DNA probes), ∼18 to 24 hr at 37◦ C. The time may be lengthened to 48 or 72 hr (as in the case with weekends), with no adverse affects. Hybridization with PNA probes require less time, at least 1 hr at room temperature. Caution, however should be taken that the slides are adequately sealed such that the probe in hybridization solution does not evaporate. Maintaining a slightly dampened slide container will help alleviate this problem. Post-hybridization washes and detection The time allotted for post-hybridization washes and detection will vary depending on the type of DNA probe used in the experiment. Directly labeled probes (DNA or PNA probes) require very little bench time since no antibody detection is required; typically they take up to 30 minutes. Probes are simply washed using high stringency washes as described in the methods. Indirectly labeled probes require incubations with antibodies (primary, secondary and/or tertiary), with the final incubation with a fluorochrome-conjugated antibody, as well as detergent washes and/or blocking steps between each antibody incubation. If the signal requires amplification, the bench time increases as each antibody incubation should last at least 30 minutes. A wash protocol using an indirectly labeled probe with signal amplification will range from 3.5 to 4 hours. Acknowledgments The authors would like to thank Elena Kolomeitz, Jana Karaskova, Paula Marrano, Ajay Pandita, Bisera Vukovic, and Zong Mei Zhang from the Department of Cellular and Molecular Biology, University Health 22.4.50 Supplement 23 Current Protocols in Cell Biology Network, Ontario Cancer Institute in Toronto, Ontario, Canada. Literature Cited Poon, S.S., Martens, U.M., Ward, R.K., Lansdorp, P.M. 1999. Telomere length measurements using digital fluorescence microscopy. Cytometry 36:267-278. Al-Romaih, K., Bayani, J., Vorobyova, J., Karaskova, J., Park, P.C., Zielenska, M., and Squire, J.A. 2003. Chromosomal instability in osteosarcoma and its association with centrosome abnormalities. Cancer Genet. Cytogenet. 144:91-99. Qian, J., Bostwick, D.G., Takahashi, S., Borell, T.J., Brown, J.A., Lieber, M.M., and Jenkins, R.B. 1996. Comparison of fluorescence in situ hybridization analysis of isolated nuclei and routine histological sections from paraffinembedded prostatic adenocarcinoma specimens. Am. J. Pathol. 149:1193-1199. Beatty, B., Mai, S., and Squire, J.A. 2002. FISH: A Practical Approach. Oxford University Press. New York. Schwarzacher, T. and Heslop-Harrison, P. 2000. Practical In Situ Hybridization. Springer-Verlag New York. Boyle, A.L., Ballard, S.G., and Ward, D.C. 1990. Differential distribution of long and short interspersed element sequences in the mouse genome: Chromosome karyotyping by fluorescence in situ hybridization. Proc. Natl. Acad. Sci. U.S.A. 87:7757-7761. Speel, E.J.M. 1999. Detection and amplification systems for sensitive, multiple-target DNA and RNA in situ hybridization: Looking inside cells with a spectrum of colors. Histochem. Cell Bio. 112:98-113. Ghadimi, B.M., Heselmeyer-Haddad, K., Auer, G., and Ried, T. 1999. Interphase cytogenetics: At the interface of genetics and morphology. Anal. Cell Pathol. 19:3-6. Giguere, V., Beatty, B., Squire, J., Copeland, N.G., and Jenkins, N.A. 1995. The orphan nuclear receptor ROR alpha (RORA) maps to a conserved region of homology on human chromosome 15q21–q22 and mouse chromosome 9. Genomics. 10:28:596-598. Henegariu, O., Heerema, N.A., Wright, L.L., BrayWard, P., Ward, D.C., and Vance, G.H. 2001. Improvements in cytogenetic slide preparation: Controlled chromosome spreading, chemical aging and gradual denaturing. Cytometry 43:101-109. Hyytinen, E., Visakorpi, T., Kallioniemi, A., Kallioniemi, O.P., and Isola, J.J. 1994. Improved technique for analysis of formalin-fixed, paraffin-embedded tumors by fluorescence in situ hybridization. Cytometry 1;16:93-99. Kolomietz, E., Al-Maghrabi, J., Brennan, S., Karaskova, J., Minkin, S., Lipton, J., and Squire, J.A. 2001. Primary chromosomal rearrangements of leukemia are frequently accompanied by extensive submicroscopic deletions and may lead to altered prognosis. Blood 97:3581-8. Speicher, M.R., Jauch, A., Walt, H., du Manoir, S., Ried, T., Jochum, W., Sulser, T., and Cremer, T. 1995. Correlation of microscopic phenotype with genotype in a formalin-fixed, paraffinembedded testicular germ cell tumor with universal DNA amplification, comparative genomic hybridization, and interphase cytogenetics. Am. J. Pathol. 146:1332-1340. Squire, J., Meurs, E.F., Chong, K.L., McMillan, N.A., Hovanessian, A.G., and Williams, B.R. 1993. Localization of the human interferoninduced, ds-RNA activated p68 kinase gene (PRKR) to chromosome 2p21–p22. Genomics 16:768-770. Squire, J.A., Thorner, P., Marrano, P., Parkinson, D., Ng, Y.K., Gerrie, B., Chilton-Macneill, S., and Zielenska, M. 1996. Identification of MYCN copy number heterogeneity by direct FISH analysis of neuroblastoma preparations. Mol. Diagn. 1:281-289. Thompson, C.T., LeBoit, P.E., Nederlof, P.M., and Gray, J.W. 1994. Thick-section fluorescence in situ hybridization on formalin-fixed, paraffinembedded archival tissue provides a histogenetic profile. Am. J. Pathol. 144:237-243. Lansdorp, P.M. 1996. Close encounters of the PNA kind. Nat. Biotechnol. 14:1653. van de Rijke, F.M., Vrolijk, H., Sloos, W., Tanke, H.J., and Raap, A.K. 1996. Sample preparation and in situ hybridization techniques for automated molecular cytogenetic analysis of white blood cells. Cytometry 24:151-157. Lichter, P., Cremer, T., Borden, J., Manuelidis, L., and Ward., D.C. 1988. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 80:224-234. Vukovic, B., Park, P.C., Al-Maghrabi, J., Beheshti, B., Sweet, J., Evans, A., Trachtenberg, J., and Squire, J. 2003. Evidence of multifocality of telomere erosion in high-grade prostatic intraepithelial neoplasia (HPIN) and concurrent carcinoma. Oncogene 22:1978-1987. Munne, S., Marquez, C., Magli, C., Morton, P., and Morrison, L. 1998. Scoring criteria for preimplantation genetic diagnosis of numerical abnormalities for chromosomes X, Y, 13, 16, 18 and 21. Mol. Hum. Reprod. 4:863-870. Martens, U.M., Zijlmans, J.M., Poon, S.S., Dragowska, W., Yui, J., Chavez, E.A., Ward, R.K., and Lansdorp., P.M. 1998. Short telomeres on human chromosome 17p. Nat. Genet. 18:76-80. Internet Resources http://www.appliedbiosystems.com The Applied Biosystems website. This company supplies PNA probes and custom orders PNA probes. http://www.cytocell.co.uk The Cytocell website. This is a commercial supplier of DNA FISH probes Cell Biology of Chromosomes and Nuclei 22.4.51 Current Protocols in Cell Biology Supplement 23 http://www.biochem.roche.com The Roche Molecular Biochemicals website. This company is a supplier of antibody and labeling reagents. http://www.vysis.com The Vysis website. This company is a supplier of commercial DNA FISH probes. Contributed by Jane Bayani and Jeremy A. Squire University of Toronto Ontario, Canada Fluorescence In Situ Hybridization (FISH) 22.4.52 Supplement 23 Current Protocols in Cell Biology